Tissue Mapping Centers
Kidney Single-Cell and Spatial Molecular Atlas Project - KIDSSMAP

Abstract
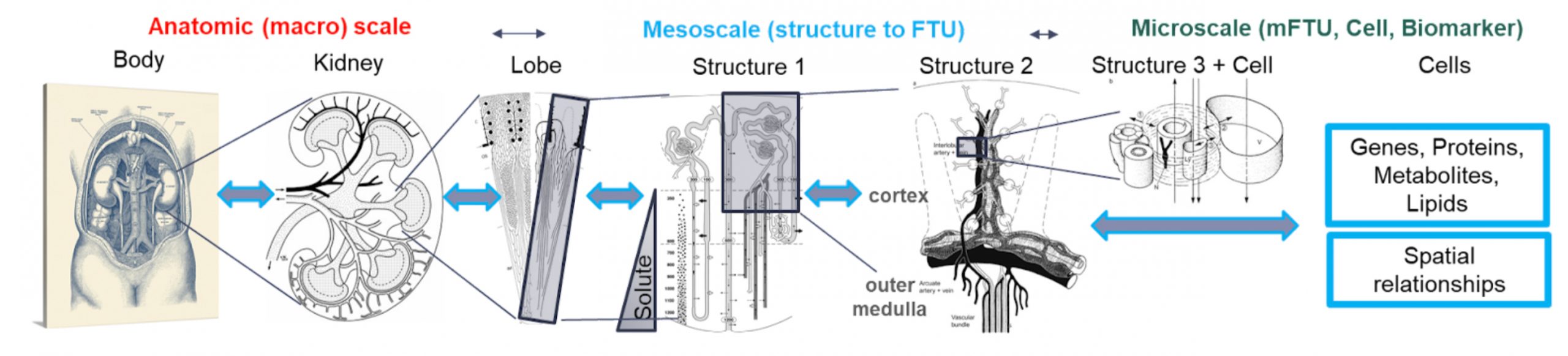
The kidney is a vital organ that maintains the composition of blood (water conservation, acid, base and electrolyte balance), has a barrier function, regulates blood pressure and erythropoiesis to enable life on land. This intricate physiology is built upon a complex anatomical, structural and molecular organization between the main functional tissue unit (FTU) of the kidney, the nephron, and surrounding niche comprising stroma, vasculature, nerves and immune cells. There is remarkable biological heterogeneity in the nephron and its niche along the corticomedullary axis with many microscopic regions working in concert to maintain kidney function. These microFTU (mFTU) affect blood filtration, solute absorption, salt and water balance. Deciphering the molecular, cellular and neighborhood diversity in mFTUs will likely provide key insights into devastating conditions such as acute, chronic and end stage kidney disease (>25 million people and >$100 billion annually) for which there are no cures.
The overarching vision of the KIDney Single cell and Spatial Molecular Atlas Project (KIDSSMAP) is to create an anatomical and molecular landscape of the human kidney that includes spatially resolved maps of the epigenome, transcriptome, proteins, metabolites, lipids and extracellular mass at a single cell resolution. These maps will be linked to construct a 3-Dimensional atlas and transform our foundational knowledge of the kidney for students(textbooks), for bioengineers to use as a guide to make mature kidneys, for biologists to understand molecular structure and function of the kidneys, to gain insights into aging, and help patients and clinician to design better drugs that target deviations from healthy states. The maps will allow integration of common cell types, genes, neighborhoods or pathways shared across organs in the body to better understand how it functions in an integrated manner. We will specifically generate high-resolution data to capture different classes of molecular moieties: RNA expression, chromatin accessibility, protein expression, lipids and metabolites across scales. Most of these assays will be done on the same tissue block and include diverse biological variables (e.g., sex, age, race). The resulting atlas will illuminate principles underlying the complex microanatomical organization of the kidney and will provide a much-needed integrated baseline for characterizing aberrant states of the cells or tissue that might arise during disease.
Fast Facts
| Project title: | Kidney Single-Cell and Spatial Molecular Atlas Project - KIDSSMAP |
| Organ specialty: | Kidney |
| PIs: | Sanjay Jain, Tarek El-Achkar |
| Co-Investigators/ Collaborators: | Joseph P. Gaut, Michael Eadon, Seth Winfree, Lingyan Shi, Kun Zhang |
| Program Managers: | Ravi Misra, PhD (OSP) and Jeanne Holden-Wiltse, MS (DAC) |
| Project Manager: | Madhurima Kaushal |
| Assay Types: | 10X Multiome (snRNA-seq, snATAC-seq), CODEX, Visium ST, SRS, CosMx (nanostring) |
| Grant number: | 1U54DK134301 |
