Transformative Technology Development
Novel Platform for Quantitative Subcellular Resolution Imaging of Human Tissues Using Mass Spectrometry

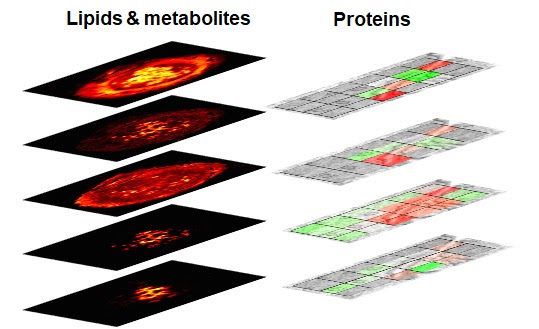
This multi-institutional TTD combines several highly innovative approaches to address challenges associated with the high-throughput, high-resolution ambient mass spectrometry imaging (MSI) of lipids, metabolites, and proteins. Lipid and metabolite imaging is performed using nanospray desorption electrospray ionization (nano-DESI), an ambient ionization technique developed by the Purdue team. Nano-DESI relies on gentle localized liquid extraction of molecules from tissue sections. Meanwhile, imaging of proteins utilizes nanoPOTS (nanodroplet Processing in One pot for Trace Samples) developed by the team from the Pacific Northwest National Laboratory (PNNL). The nanoPOTs MSI platform combines laser capture microdissection, sample preparation, nanoLC-MS/MS, and an open-source bioinformatic tool (Trelliscope) for data processing and visualization. The throughput of both experimental techniques is enhanced using Deep Learning Approach for Dynamic Sparse Sampling (DLADS) developed by the Marquette team.
They have developed a novel microfluidic probe for nano-DESI MSI experiments optimized for robustness, ease of fabrication, and spatial resolution. The probe provides stable signals from different types of tissues and is capable of imaging lipids and metabolites with high spatial resolution and throughput (https://onlinelibrary.wiley.com/doi/full/10.1002/ange.202006531). The PNNL team has demonstrated that the nanoPOTS MSI platform can quantitatively generate cell-type-specific images for >2,000 proteins at 100-µm spatial resolution across 12-µm thick tissue sections using a label free quantitation approach (https://www.nature.com/articles/s41467-019-13858-z). To increase analysis throughput, detection sensitivity, and improve spatial resolution, without compromising proteome coverage, the PNNL team is advancing the nanoPOTS MSI workflow by incorporating tandem mass tags (TMT) labeling. DLADS developed within this project has been used to further improve the experimental throughput. DLADS dynamically determines optimal sampling locations where high-fidelity reconstruction is achieved. Inter-pixel relationship among on-the-fly samples is estimated by the convolutional neural networks trained on off-line training datasets so that measurements are focused on highly informative locations (e.g., gradient). DLADS showed 6-fold throughput improvement in nano-DESI simulation by significantly reducing the number of required measurements.
These developments will be applicable to different types of human tissues and will transform quantitative molecular imaging of multiple classes of biomolecules in tissue sections. Although the capabilities of the new imaging platform will be demonstrated using non-diseased tissue, these developments will be broadly applicable to scientific problems associated with understanding health and disease.
Fast Facts
| Project title: | Novel Platform for Quantitative Subcellular Resolution Imaging of Human Tissues Using Mass Spectrometry |
| Organ specialty: | Kidney, Heart, Pancreas |
| PI: | Julia Laskin |
| Co-Investigators: | Purdue: Xiangtang Li, Hang Hu, Manxi Yang PNNL: Kristin Burnum-Johnson, Ying Zhu, Paul Piehowski, Lisa Bramer Marquette: Dong Hye Ye, David Helminiak |
| Project Manager: | Manxi Yang |
| Assay types: | NanoDESI; nanoPOTS |
| Grant number: | 1UG3HL145593-01 |
| Learn more: | Visit the Laskin lab website, and follow them on Twitter. |
