HuBMAP Integration, Visualization & Engagement
Tools Component
Comprehensive, Flexible and Fair Tools for the HuBMAP HIVE

This group works to develop computational methods and tools to enable the analysis, integration and use of HuBMAP collected data by consortium participants, the international research community and the large software and method development community.
There are three key directions for the Tools Component: (1) Pipelines for uniform processing of all HuBMAP generated data. Current pipelines include those for single cell sequencing data (including scRNA-Seq and ATAC-Seq), pipelines for HuBMAP imaging data (including segmentation and stitching), pipelines for spatial transcriptomics and spatial proteomics data (including codex, seqfish etc.) and pipelines for downstream analysis of all image based HuBMAP data. (2) Community involvement: Development of repositories, indexing, APIs, interfaces and schemas to enable the hosting, discovery and integration of processed data and its use by the community. (3) Innovative tool development including new methods for the analysis of single cell sequencing data (cell type assignments, cell-cell interactions), new methods for segmentation of various imaging datasets, new methods for visualization of large high throughput datasets and more.
All tools created by the group including pipelines and APIs are publicly available. The group commits to ensuring its tools are findable, accessible, interoperable and reusable (FAIR) and has in place plans to address issues related to validation of tool accuracy and usefulness, data privacy, and reproducibility. The Tools component works very closely with other HIVE components to disseminate the tools and to connect with both the developers and the international research communities.
Explore our analysis tools:
- scRNA-seq pipeline: Salmon, Scanpy, scVelo
- scATAC-seq pipeline: SnapTools, SnapATAC, chromVAR
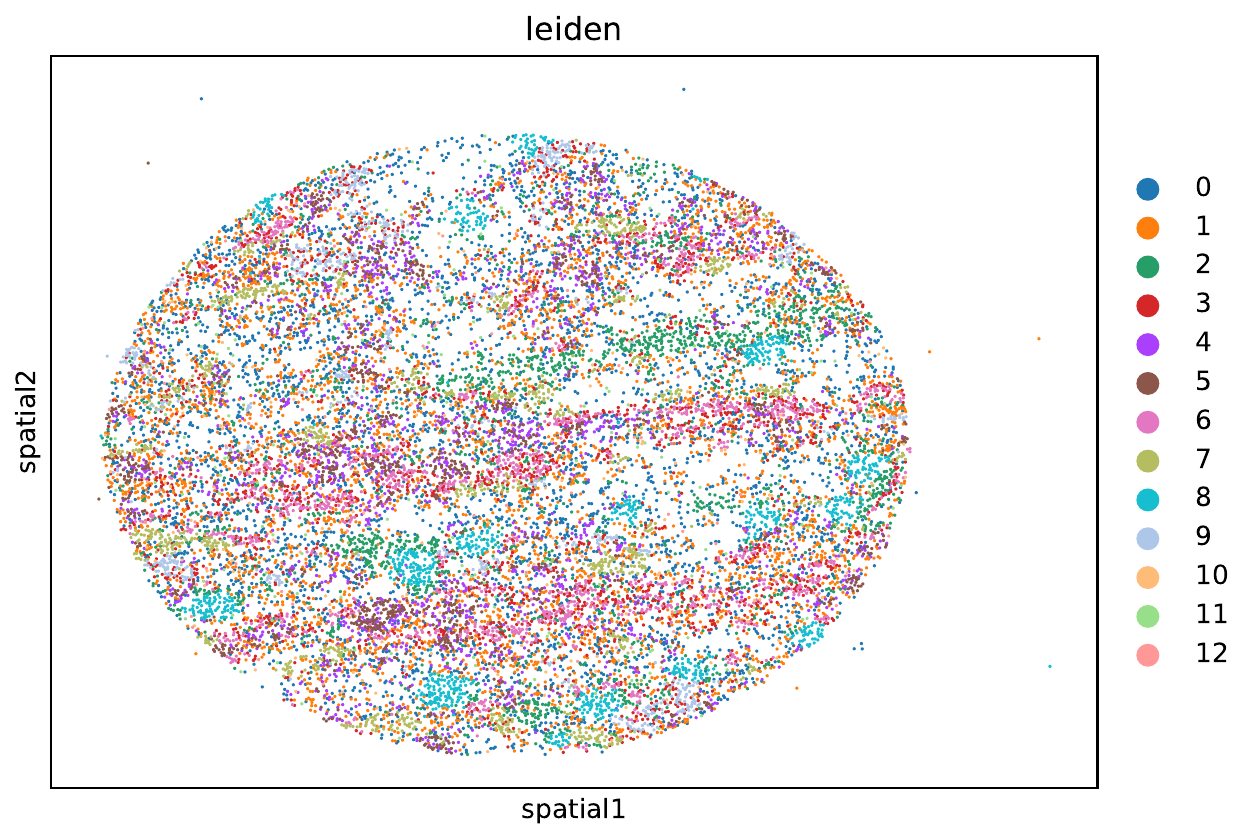
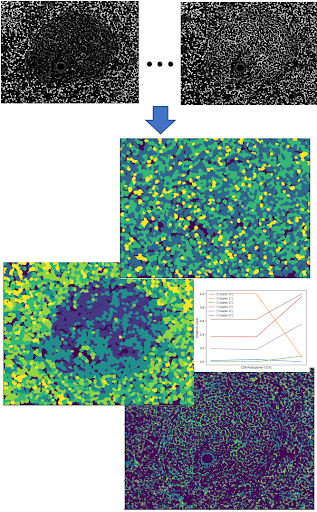
- SPRM
- CODEX pipeline: Cytokit, SPRM
- Cells API backend service (Python client, Javascript client, cell ID generation library)
- IMS + MxIF pipeline
Fast Facts
| Project title: | Comprehensive, Flexible and Fair Tools for the HuBMAP HIVE |
| PI: | Matt Ruffalo, Carnegie Mellon University |
| Co-Investigators: | Benedict Paten, University of California, Santa Cruz; Sarah Teichmann, Wellcome Sanger Institute |
| Project Manager: | Vladimir Kiselev, Wellcome Sanger Institute; Allyson (Ally) Ricarte, Carnegie Mellon University |
| Grant number: | 1OT2OD026682-01 |
| Learn more: | Visit the lab website. |
