About HuBMAP Data
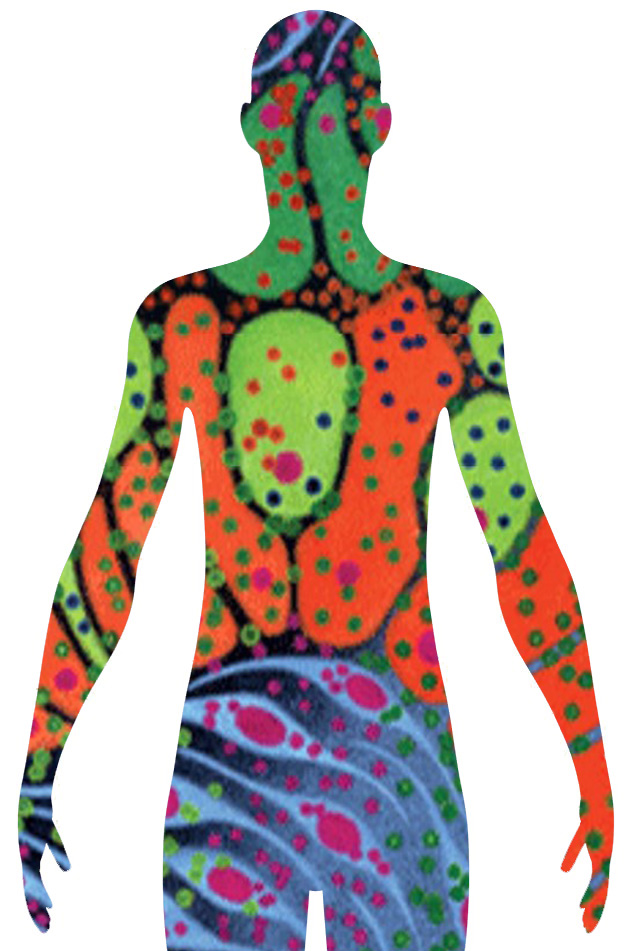
The focus of HuBMAP is understanding the intrinsic intra-, inter-, and extra- cellular biomolecular distribution in human tissue. HuBMAP will focus on fresh, fixed, or frozen healthy human tissue using in situ and dissociative techniques that have high-spatial resolution.
The HuBMAP Data Portal aims to be an open map of the human body at the cellular level. These tools and maps are openly available, to accelerate understanding of the relationships between cell and tissue organization and function and human health.
The Human Reference Atlas is a comprehensive, high-resolution, three-dimensional atlas of all the cells in the healthy human body. The Human Reference Atlas provides standard terminologies and data structures for describing specimens, biological structures, and spatial positions linked to existing ontologies.
Explore the Data
HuBMAP-developed tools like the Human Reference Atlas, Vitessce, and Azimuth allow visual exploration of HuBMAP tissues and cells. You can also search the data in the Data Portal directly, and download data for your use.
Search the Data
Anyone can access the Data Portal without logging in to browse, download metadata, and visualize public data.
Viewers can browse by donors, samples, datasets, and collections, and refine their search by organ type, specimen type, assay type, donor information, affiliation (who produced the data), and more.
Download Data
While an account is not necessary to access and view datasets within the Data Portal, viewers who log in are able to download processed data.
Login to the Data Portal is handled through Globus. Login with your institutional or eRACommons credentials for anonymous access to file downloads.
Vitessce
Vitessce (Visual integration tool for exploration of spatial single cell experiments) enables the exploration of spatially-resolved, integrated single-cell datasets, witj reusable components such as a scatterplot, spatial+imaging plot, genome browser tracks, statistical plots, and controller components. An interactive visualization tool, Vitessce is available for datasets that have been processed through HuBMAP HIVE computing hardware and pipelines.
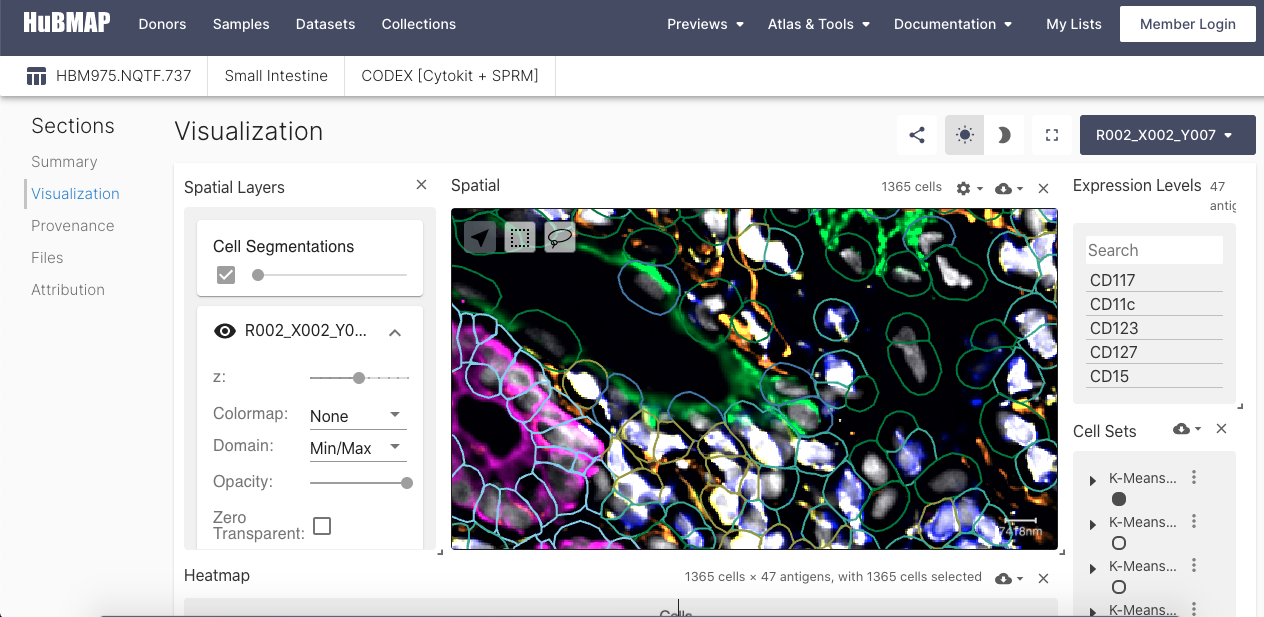
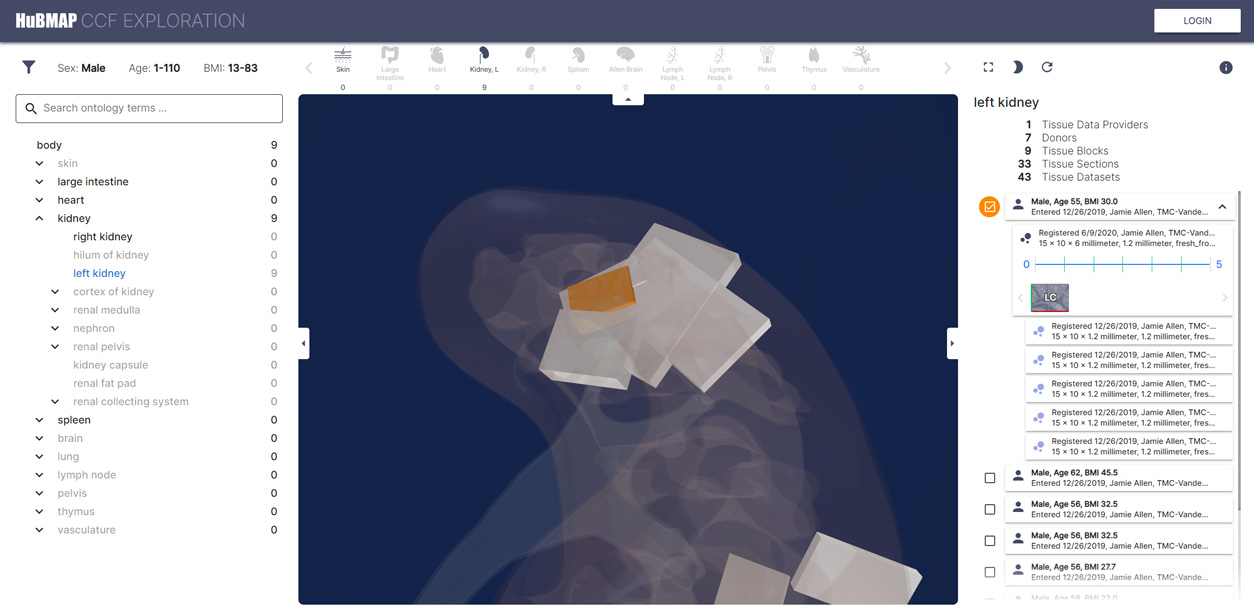
Human Reference Atlas Exploration User Interface
A Human Reference Atlas (HRA) is a comprehensive, high-resolution, three-dimensional atlas of all the cells in the human body. The HRA provides standard terminologies and data structures for describing specimens, biological structures, and spatial positions linked to existing ontologies.
The HRA consists of several tools. One of them, the Exploration User Interface is available in the HUBMAP data portal. To see them all, visit the HRA site.
About the Data
HuBMAP is transparent about the origin of its data, how the data is processed, and the quality control processes used to ensure its integrity.
Organs
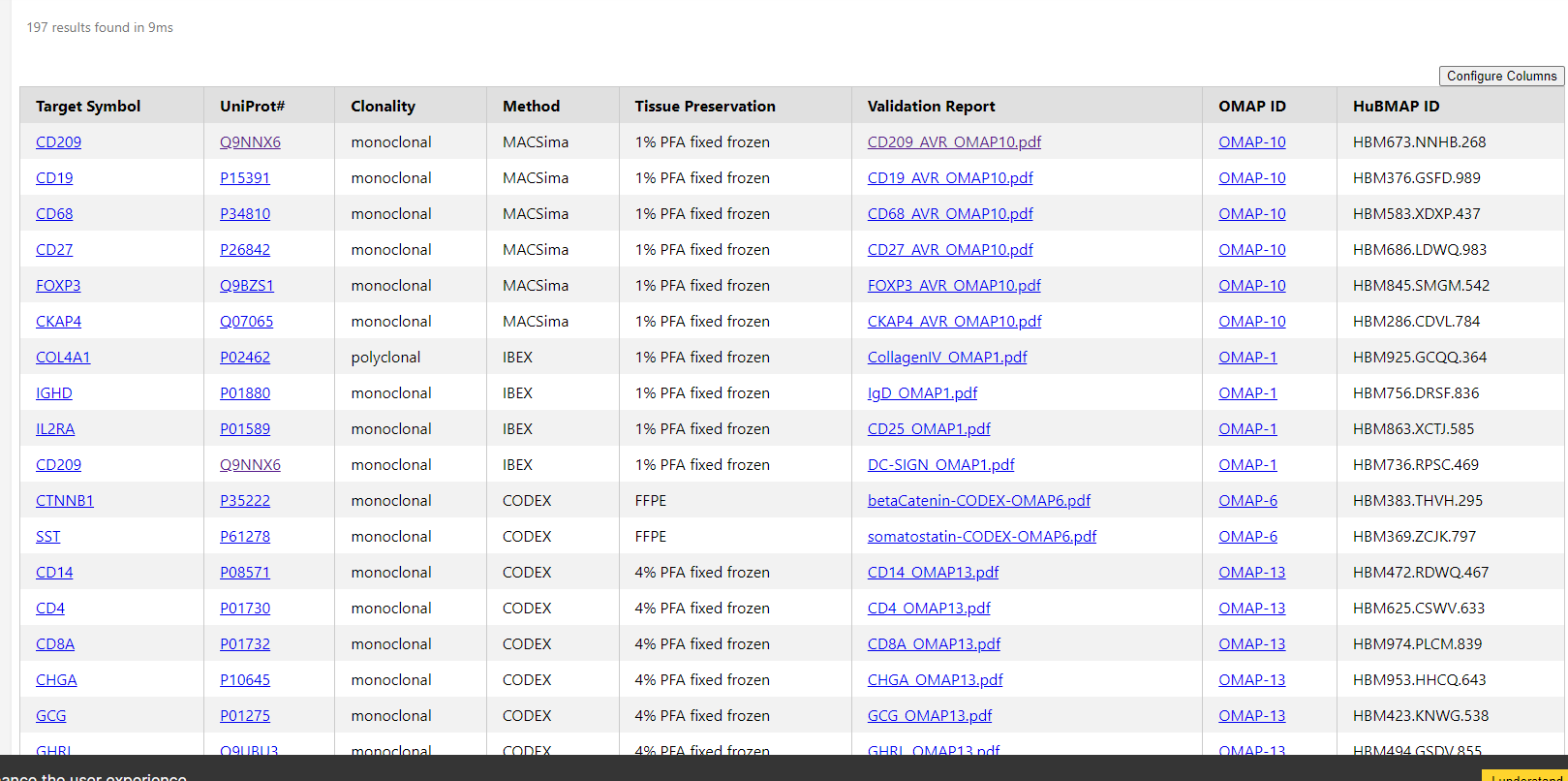
HuBMAP data encompasses over 2300 data sets spanning more than 30 organs. Specimen types (fresh frozen tissue section, FFPE, single cell cryopreserved, etc.) are associated with the organs and can be used to filter during exploration.
The HuBMAP data portal contains an overview page for each organ. This overview includes information such as the assays used on the tissue, visuals of the organ tissue in the Human Reference Atlas, and links to specific tissue samples. Drilling down to specific samples will reveal details on metadata, processing pipelines, and links to download public data.
Click on an organ to see it in our data portal.
HuBMAP Data comprises healthy (normal, non-diseased) tissue from over 200 donors. The data for sex, race, and ethnicity provided are those pulled from the electronic medical records available and do not reflect the breadth of human identity and experience. We hope to expand the diversity of our donors in future data releases.