HuBMAP Publications
As of May 2024, HuBMAP researchers have published more than 330 scientific papers, presenting advances in software and experimental technologies, and sharing progress in creating maps of healthy human tissues at the cellular level.
We feature a small selection of those publications below, but you can view all of our publications in Google Scholar to learn more about the advances we are making towards creating maps of individual cells of the human body in two and three dimensions.
Featured Publications
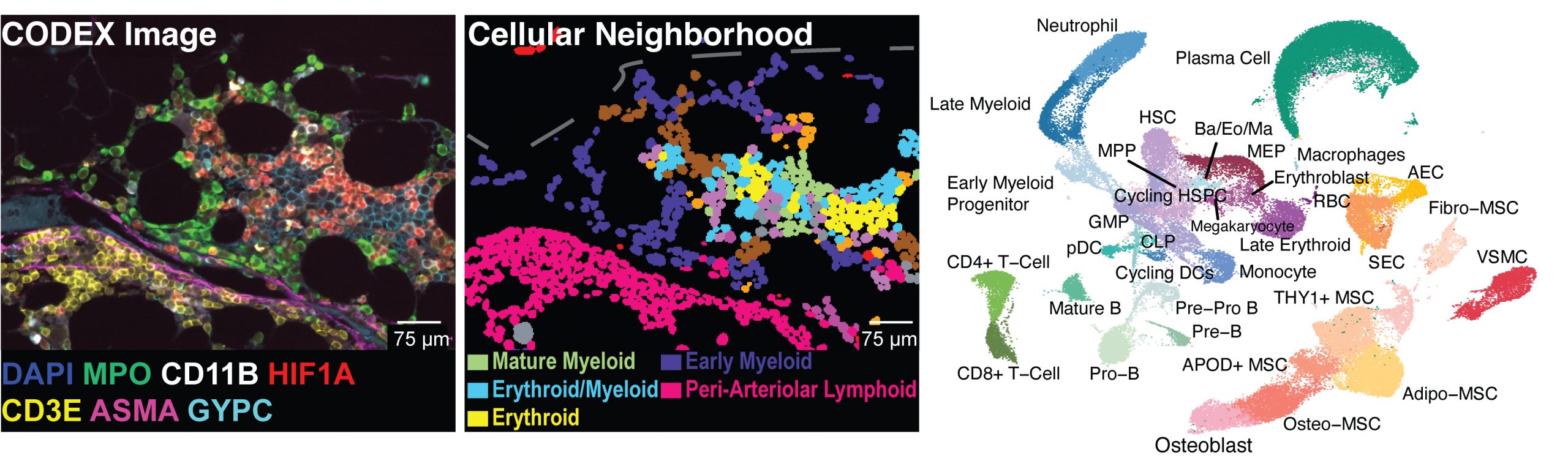
Mapping the cellular biogeography of human bone marrow niches using single-cell transcriptomics and proteomic imaging
Bandyopadhyay S, Duffy MP, Ahn KJ, Sussman JH, Pang M, Smith D, Duncan G, Zhang I, Huang J, Lin Y, Xiong B, Imtiaz T, Chen CH, Thadi A, Chen C, Xu J, Reichart M, Martinez Z, Diorio C, Chen C, Pillai V, Snaith O, Oldridge D, Bhattacharyya S, Maillard I, Carroll M, Nelson C, Qin L, Tan K.
Abstract
Non-hematopoietic cells are essential contributors to hematopoiesis. However, heterogeneity and spatial organization of these cells in human bone marrow remain largely uncharacterized. We used single-cell RNA sequencing (scRNA-seq) to profile 29,325 non-hematopoietic cells and discovered nine transcriptionally distinct subtypes. We simultaneously profiled 53,417 hematopoietic cells and predicted their interactions with non-hematopoietic subsets. We employed co-detection by indexing (CODEX) to spatially profile over 1.2 million cells. We integrated scRNA-seq and CODEX data to link predicted cellular signaling with spatial proximity. Our analysis revealed a hyperoxygenated arterio-endosteal neighborhood for early myelopoiesis, and an adipocytic localization for early hematopoietic stem and progenitor cells (HSPCs). We used our CODEX atlas to annotate new images and uncovered mesenchymal stromal cell (MSC) expansion and spatial neighborhoods co-enriched for leukemic blasts and MSCs in acute myeloid leukemia (AML) patient samples. This spatially resolved, multiomic atlas of human bone marrow provides a reference for investigation of cellular interactions that drive hematopoiesis.
Additional Resources
Software and source code created for this study are publicly available and can be obtained at GitHub at https://github.com/tanlabcode/spatial-bonemarrow-atlas. Single-cell RNA-seq data can be downloaded from NCBI Gene Expression Omnibus (GEO) database with the accession number GSE253355 . This includes the raw sequencing reads, count matrices, and annotated Seurat object.
Primary imaging data and processed CODEX data are publicly available and can be downloaded from FigShare at: https://doi.org/10.25452/figshare.plus.c.7174914.
This includes full multiplexed images for the normal bone marrow atlas, AML, NSM samples, corresponding cell segmentation masks and H&E images as well as annotated Seurat objects which contain unnormalized and normalized marker expression values for each cell.
Both scRNA-seq and CODEX data are publicly available to explore interactively through Vitessce widgets hosted at https://cscb.research.chop.edu/index.php/bm-data.
Corresponding author
Kai Tan
Contributing groups
Grant support
NIH Award Nos. U54HL156090 and U54HL165442
Unsupervised and supervised discovery of tissue cellular neighborhoods from cell phenotypes
Hu Y, Rong J, Xu Y, Xie R, Peng J, Gao L, Tan K
Abstract
It is poorly understood how different cells in a tissue organize themselves to support tissue functions. We describe the CytoCommunity algorithm for the identification of tissue cellular neighborhoods (TCNs) based on cell phenotypes and their spatial distributions. CytoCommunity learns a mapping directly from the cell phenotype space to the TCN space using a graph neural network model without intermediate clustering of cell embeddings. By leveraging graph pooling, CytoCommunity enables de novo identification of condition-specific and predictive TCNs under the supervision of sample labels. Using several types of spatial omics data, we demonstrate that CytoCommunity can identify TCNs of variable sizes with substantial improvement over existing methods. By analyzing risk-stratified colorectal and breast cancer data, CytoCommunity revealed new granulocyte-enriched and cancer-associated fibroblast-enriched TCNs specific to high-risk tumors and altered interactions between neoplastic and immune or stromal cells within and between TCNs. CytoCommunity can perform unsupervised and supervised analyses of spatial omics maps and enable the discovery of condition-specific cell-cell communication patterns across spatial scales.
Corresponding authors
Yuxuan Hu and Kai Tan
Contributing groups
Grant support
NIH Award Nos. U54HL156090 and U54HL165442
2023 Nature Publications
Thirteen scientific papers arising from HuBMAP research are featured in the July 2023 issues of the Nature family of journals, as part of a larger collection of HuBMAP publications. We are proud to highlight those publications here.
You can peruse the entire collection at https://hubmapconsortium.org/nature-package.
Advances and Prospects for the Human Biomolecular Atlas Program
Jain S, Pei L, Spraggins JM, Angelo M, Carson JP, Gehlenborg N, Ginty F, Gonçalves JP, Hagood JS, Hickey JW, Kelleher NL, Laurent LC, Lin S, Lin Y, Liu H, Naba A, Nakayasu ES, Qian WJ, Radtke A, Robson P, Stockwell BR, Van de Plas R, Vlachos IS, Zhou M; HuBMAP Consortium; Börner K, Snyder MP
Published in Nature Cell Biology, July 2023.
Abstract
The Human BioMolecular Atlas Program (HuBMAP) aims to create a multi-scale spatial atlas of the healthy human body at single-cell resolution by applying advanced technologies and disseminating resources to the community. As the HuBMAP moves past its first phase, creating ontologies, protocols and pipelines, this Perspective introduces the production phase: the generation of reference spatial maps of functional tissue units across many organs from diverse populations and the creation of mapping tools and infrastructure to advance biomedical research.
Grant support
NIH Award Nos. U54 HL165442, U01 HL166058, U54 DK134301, OT2 OD033753, U54 EY032442, U54 DK120058, U54 DK134302, OT2 OD033756, OT2 OD026671, UH3 CA246635, UG3 CA256967, U54 HG010426, UH3 CA246633, U54 HD104393, U54 DK127823, OT2 OD033758, UH3 CA246594, U54 EY032442, U54 DK134302, U54 HL145611, U01 HG012680, U54 DK120058, U54 EY032442, U54 DK134302, UG3 CA256959, and U54 HL165440
Corresponding Authors
Sanjay Jain
Liming Pei
Jeffrey Spraggins
Katy Börner
Michael P. Snyder
Contributing groups
An atlas of healthy and injured cell states and niches in the human kidney
Lake BB, Menon R, Winfree S, Hu Q, Ferreira RM, Kalhor K, Barwinska D, Otto EA, Ferkowicz M, Diep D, Plongthongkum N, Knoten A, Urata S, Mariani LH, Naik AS, Eddy S, Zhang B, Wu Y, Salamon D, Williams JC, Wang X, Balderrama KS, Hoover PJ, Murray E, Marshall JL, Noel T, Vijayan A, Hartman A, Chen F, Waikar SS, Rosas SE, Wilson FP, Palevsky PM, Kiryluk K, Sedor JR, Toto RD, Parikh CR, Kim EH, Satija R, Greka A, Macosko EZ, Kharchenko PV, Gaut JP, Hodgin JB; KPMP Consortium; Eadon MT, Dagher PC, El-Achkar TM, Zhang K, Kretzler M, Jain S.
Published in Nature, July 2023.
Abstract
Understanding kidney disease relies on defining the complexity of cell types and states, their associated molecular profiles and interactions within tissue neighbourhoods1. Here we applied multiple single-cell and single-nucleus assays (>400,000 nuclei or cells) and spatial imaging technologies to a broad spectrum of healthy reference kidneys (45 donors) and diseased kidneys (48 patients). This has provided a high-resolution cellular atlas of 51 main cell types, which include rare and previously undescribed cell populations. The multi-omic approach provides detailed transcriptomic profiles, regulatory factors and spatial localizations spanning the entire kidney. We also define 28 cellular states across nephron segments and interstitium that were altered in kidney injury, encompassing cycling, adaptive (successful or maladaptive repair), transitioning and degenerative states. Molecular signatures permitted the localization of these states within injury neighbourhoods using spatial transcriptomics, while large-scale 3D imaging analysis (around 1.2 million neighbourhoods) provided corresponding linkages to active immune responses. These analyses defined biological pathways that are relevant to injury time-course and niches, including signatures underlying epithelial repair that predicted maladaptive states associated with a decline in kidney function. This integrated multimodal spatial cell atlas of healthy and diseased human kidneys represents a comprehensive benchmark of cellular states, neighbourhoods, outcome-associated signatures and publicly available interactive visualizations.
Additional Resources
Corresponding Authors
Sanjay Jain
Grant support
NIH Award Nos. U54 DK134301, OT2 OD033753, U54 HL145608, and UH3 CA246632
Organization of the Human Intestine at Single Cell Resolution
Hickey JW, Becker WR, Nevins SA, Horning A, Perez AE, Zhu C, Zhu B, Wei B, Chiu R, Chen DC, Cotter DL, Esplin ED, Weimer AK, Caraccio C, Venkataraaman V, Schürch CM, Black S, Brbić M, Cao K, Chen S, Zhang W, Monte E, Zhang NR, Ma Z, Leskovec J, Zhang Z, Lin S, Longacre T, Plevritis SK, Lin Y, Nolan GP, Greenleaf WJ, Snyder M.
Published in Nature, July 2023.
Abstract
The intestine is a complex organ that promotes digestion, extracts nutrients, participates in immune surveillance, maintains critical symbiotic relationships with microbiota and affects overall health1. The intesting has a length of over nine metres, along which there are differences in structure and function2. The localization of individual cell types, cell type development trajectories and detailed cell transcriptional programs probably drive these differences in function. Here, to better understand these differences, we evaluated the organization of single cells using multiplexed imaging and single-nucleus RNA and open chromatin assays across eight different intestinal sites from nine donors. Through systematic analyses, we find cell compositions that differ substantially across regions of the intestine and demonstrate the complexity of epithelial subtypes, and find that the same cell types are organized into distinct neighbourhoods and communities, highlighting distinct immunological niches that are present in the intestine. We also map gene regulatory differences in these cells that are suggestive of a regulatory differentiation cascade, and associate intestinal disease heritability with specific cell types. These results describe the complexity of the cell composition, regulation and organization for this organ, and serve as an important reference map for understanding human biology and disease.
Additional Resources
Corresponding Authors
Michael Snyder
Contributing groups
Grant support
NIH Award No. U54 HG012723
A spatially resolved timeline of the human maternal-fetal interface
Greenbaum S, Averbukh I, Soon E, Rizzuto G, Baranski A, Greenwald NF, Kagel A, Bosse M, Jaswa EG, Khair Z, Kwok S, Warshawsky S, Piyadasa H, Goldston M, Spence A, Miller G, Schwartz M, Graf W, Van Valen D, Winn VD, Hollmann T, Keren L, van de Rijn M, Angelo M.
Published in Nature, July 2023.
Abstract
Beginning in the first trimester, fetally derived extravillous trophoblasts (EVTs) invade the uterus and remodel its spiral arteries, transforming them into large, dilated blood vessels. Several mechanisms have been proposed to explain how EVTs coordinate with the maternal decidua to promote a tissue microenvironment conducive to spiral artery remodelling (SAR)1-3. However, it remains a matter of debate regarding which immune and stromal cells participate in these interactions and how this evolves with respect to gestational age. Here we used a multiomics approach, combining the strengths of spatial proteomics and transcriptomics, to construct a spatiotemporal atlas of the human maternal-fetal interface in the first half of pregnancy. We used multiplexed ion beam imaging by time-of-flight and a 37-plex antibody panel to analyse around 500,000 cells and 588 arteries within intact decidua from 66 individuals between 6 and 20 weeks of gestation, integrating this dataset with co-registered transcriptomics profiles. Gestational age substantially influenced the frequency of maternal immune and stromal cells, with tolerogenic subsets expressing CD206, CD163, TIM-3, galectin-9 and IDO-1 becoming increasingly enriched and colocalized at later time points. By contrast, SAR progression preferentially correlated with EVT invasion and was transcriptionally defined by 78 gene ontology pathways exhibiting distinct monotonic and biphasic trends. Last, we developed an integrated model of SAR whereby invasion is accompanied by the upregulation of pro-angiogenic, immunoregulatory EVT programmes that promote interactions with the vascular endothelium while avoiding the activation of maternal immune cells.
Additional Resources
Corresponding Authors
Robert Michael Angelo
Grant support
NIH Award Nos. U54 HL165445 and UH3 CA246633
3D reconstruction of skin and spatial mapping of immune cell density, vascular distance and effects of sun exposure and aging
Ghose S, Ju Y, McDonough E, Ho J, Karunamurthy A, Chadwick C, Cho S, Rose R, Corwin A, Surrette C, Martinez J, Williams E, Sood A, Al-Kofahi Y, Falo LD Jr, Börner K, Ginty F.
Published in Communications Biology, July 2023.
Abstract
Mapping the human body at single cell resolution in three dimensions (3D) is important for understanding cellular interactions in context of tissue and organ organization. 2D spatial cell analysis in a single tissue section may be limited by cell numbers and histology. Here we show a workflow for 3D reconstruction of multiplexed sequential tissue sections: MATRICS-A (Multiplexed Image Three-D Reconstruction and Integrated Cell Spatial - Analysis). We demonstrate MATRICS-A in 26 serial sections of fixed skin (stained with 18 biomarkers) from 12 donors aged between 32–72 years. Comparing the 3D reconstructed cellular data with the 2D data, we show significantly shorter distances between immune cells and vascular endothelial cells (56 µm in 3D vs 108 µm in 2D). We also show 10–70% more T cells (total) within 30 µm of a neighboring T helper cell in 3D vs 2D. Distances of p53, DDB2 and Ki67 positive cells to the skin surface were consistent across all ages/sun exposure and largely localized to the lower stratum basale layer of the epidermis. MATRICS-A provides a framework for analysis of 3D spatial cell relationships in healthy and aging organs and could be further extended to diseased organs.
Additional Resources
Datasets in the HuBMAP data portal:
- Region 1, dataset HBM732.FZVZ.656
- Region 2, dataset HBM747.SPWK.779
- Region 3, dataset HBM398.NCVN.256
- Region 4, dataset HBM746.VTDZ.959
- Region 5, HBM875.SBHJ.939
- Region 6, dataset HBM867.NMXL.794
- Region 7, dataset HBM666.JCGS.862
- Region 8, dataset HBM592.JGSQ.253
- Region 9, dataset HBM494.XDQW.356
- Region 10, dataset HBM238.ZKPC.934
- Region 11, dataset HBM975.FVCG.922
- Region 12, dataset HBM674.XQFQ.364
Corresponding Authors
Fiona Ginty
Grant support
NIH Award Nos. U54 AR081775, OT2 OD026671, and OT2 OD033756
A spatially anchored transcriptomic atlas of the human kidney papilla identifies significant immune injury in patients with stone disease
Canela VH, Bowen WS, Ferreira RM, Syed F, Lingeman JE, Sabo AR, Barwinska D, Winfree S, Lake BB, Cheng YH, Gaut JP, Ferkowicz M, LaFavers KA, Zhang K, Coe FL, Worcester E; Kidney Precision Medicine Project; Jain S, Eadon MT, Williams JC Jr, El-Achkar TM.
Published in Nature Communications, July 2023.
Abstract
Kidney stone disease causes significant morbidity and increases health care utilization. In this work, we decipher the cellular and molecular niche of the human renal papilla in patients with calcium oxalate (CaOx) stone disease and healthy subjects. In addition to identifying cell types important in papillary physiology, we characterize collecting duct cell subtypes and an undifferentiated epithelial cell type that was more prevalent in stone patients. Despite the focal nature of mineral deposition in nephrolithiasis, we uncover a global injury signature characterized by immune activation, oxidative stress and extracellular matrix remodeling. We also identify the association of MMP7 and MMP9 expression with stone disease and mineral deposition, respectively. MMP7 and MMP9 are significantly increased in the urine of patients with CaOx stone disease, and their levels correlate with disease activity. Our results define the spatial molecular landscape and specific pathways contributing to stone-mediated injury in the human papilla and identify associated urinary biomarkers.
Additional Resources
- Donor, sample and dataset information on the HuBMAP data portal for donor HBM984.MHHN.738
- Donor, sample and dataset information on the HuBMAP data portal for donor HBM375.PJSC.682
- Donor, sample and dataset information on the HuBMAP data portal for donor HBM994.SGLP.263
- Donor, sample and dataset information on the HuBMAP data portal for donor HBM825.GBHD.667
- Donor, sample and dataset information on the HuBMAP data portal for donor HBM833.LBTG.876
- Donor, sample and dataset information on the HuBMAP data portal for donor HBM277.XCLT.589
- Donor, sample and dataset information on the HuBMAP data portal for donor HBM773.MKPP.732
- Dataset SuperSeries GSE183279 in the GEO repository
- Dataset SuperSeries GSE231630 in the GEO repository
- Supporting data and analysis in Zenodo
Corresponding Authors
Sanjay Jain and Tarek El-Achkar
Grant support
NIH Award Nos. U54 DK134301, U54 HL145608, and OT2 OD033753
Anatomical structures, cell types, and biomarkers of the healthy human blood vasculature
Boppana A, Lee S, Malhotra R, Halushka M, Gustilo KS, Quardokus E, Herr II BW, Börner K, Weber GM.
Published in Scientific Data, July 2023.
Abstract
More than 150 scientists from 17 consortia are collaborating on an international project to build a Human Reference Atlas, which maps all 37 trillion cells in the healthy adult human body. The initial release of this atlas provided hierarchical lists of the anatomical structures, cell types, and biomarkers in 11 organs. Here, we describe the methods we used as part of this initiative to build the first open, computer-readable, and comprehensive database of the adult human blood vasculature, called the Human Reference Atlas-Vasculature Common Coordinate Framework (HRA-VCCF). It includes 993 vessels and their branching connections, 10 cell types, and 10 biomarkers. With this paper we are releasing additional details on vessel types and subtypes, branching sequence, anastomoses, portal systems, microvasculature, functional tissue units, mappings to regions vessels supply or drain, geometric properties of vessels, and links to 3D reference objects. Future versions will add variants and connections to the lymph vasculature; and, it will iteratively expand and improve the database as additional experimental data become available through the participating consortia.
Additional Resources
Corresponding Authors
Griffin Weber and Katy Börner
Contributing groups
Grant support
NIH Award Nos. OT2 OD026671 and OT2 OD033756
Organ Mapping Antibody Panels (OMAPs): A community resource for standardized multiplexed tissue imaging
Quardokus EM, Saunders DC, McDonough E, Hickey JW, Werlein C, Surrette C, Rajbhandari P, Casals AM, Tian H, Lowery L, Neumann EK, Björklund F, Neelakantan TV, Croteau J, Wiblin AE, Fisher J, Livengood AJ, Dowell KG, Silverstein JC, Spraggins JM, Pryhuber GS, Deutsch G, Ginty F, Nolan GP, Melov S, Jonigk D, Caldwell MA, Vlachos IS, Muller W, Gehlenborg N, Stockwell BR, Lundberg E, Snyder MP, Germain RN, Camarillo JM, Kelleher NL, Börner K, Radtke AJ
Published in Nature Methods, July 2023.
Abstract
Multiplexed antibody-based imaging enables the detailed characterization of molecular and cellular organization in tissues. Advances in the field now allow high-parameter data collection (>60 targets); however, considerable expertise and capital are needed to construct the antibody panels employed by these methods. Organ mapping antibody panels are community-validated resources that save time and money, increase reproducibility, accelerate discovery and support the construction of a Human Reference Atlas.
Additional Resources
Corresponding Authors
Andrea Radtke
Contributing groups
- HIVE Mapping Component, Indiana University
- HIVE Infrastructure and Engagement Component, Pittsburgh Supercomputing Center and University of Pittsburgh
- HIVE Tools Component, Harvard Medical School
- Rapid Technology Implementation, General Electric
- Rapid Technology Implementation, Northwestern University
- Tissue Mapping Center, Vanderbilt University (Kidney)
- Tissue Mapping Center, Vanderbilt University (Pancreas and Eye)
- Tissue Mapping Center, Stanford University
- Tissue Mapping Center, Beth Israel Deaconess Medical Center
- Tissue Mapping Center, University of California San Diego
- Transformative Technology Development, Harvard University
- Transformative Technology Development, Columbia University and Pennsylvania State University
Grant support
NIH Award Nos. UG3 CA256962, OT2 OD026671, OT2 OD033756, OT2 OD026675, OT2 OD033758, U54 DK134302, U54 EY032442, U54 HG010426, U54 HL165440, U54 HL145608, UG3 HL145600, UH3 CA246594, UH3 CA246635, and UH3 CA246635
Segmentation of human functional tissue units in support of a Human Reference Atlas
Jain Y, Godwin LL, Ju Y, Sood N, Quardokus EM, Bueckle A, Longacre T, Horning A, Lin Y, Esplin ED, Hickey JW, Snyder MP, Patterson NH, Spraggins JM, Börner K.
Published in Commun. Biol., July 2023.
Abstract
The Human BioMolecular Atlas Program (HuBMAP) aims to compile a Human Reference Atlas (HRA) for the healthy adult body at the cellular level. Functional tissue units (FTUs), relevant for HRA construction, are of pathobiological significance. Manual segmentation of FTUs does not scale; highly accurate and performant, open-source machine-learning algorithms are needed. We designed and hosted a Kaggle competition that focused on development of such algorithms and 1200 teams from 60 countries participated. We present the competition outcomes and an expanded analysis of the winning algorithms on additional kidney and colon tissue data, and conduct a pilot study to understand spatial location and density of FTUs across the kidney. The top algorithm from the competition, Tom, outperforms other algorithms in the expanded study, while using fewer computational resources. Tom was added to the HuBMAP infrastructure to run kidney FTU segmentation at scale-showcasing the value of Kaggle competitions for advancing research.
Additional Resources
- "Hacking the Kidney" 2021 Kaggle Competition dataset in the HuBMAP data portal - Collection ID HBM925.SGXL.596
- Five winning algorithms from the 2021 Kaggle Competition
- Data for this paper
- Trained models for this paper
- Code for this paper
- Competition website
- Publication page on the HuBMAP data portal
Corresponding Authors
Katy Börner
Grant support
NIH Award Nos. OT2 OD026671, U54 EY032442, U54 HG010426, and U54 DK120058
Specimen, biological structure, and spatial ontologies in support of a Human Reference Atlas
Herr BW 2nd, Hardi J, Quardokus EM, Bueckle A, Chen L, Wang F, Caron AR, Osumi-Sutherland D, Musen MA, Börner K.
Published in Sci Data, March 2023.
Abstract
The Human Reference Atlas (HRA) is defined as a comprehensive, three-dimensional (3D) atlas of all the cells in the healthy human body. It is compiled by an international team of experts who develop standard terminologies that they link to 3D reference objects, describing anatomical structures. The third HRA release (v1.2) covers spatial reference data and ontology annotations for 26 organs. Experts access the HRA annotations via spreadsheets and view reference object models in 3D editing tools. This paper introduces the Common Coordinate Framework (CCF) Ontology v2.0.1 that interlinks specimen, biological structure, and spatial data, together with the CCF API that makes the HRA programmatically accessible and interoperable with Linked Open Data (LOD). We detail how real-world user needs and experimental data guide CCF Ontology design and implementation, present CCF Ontology classes and properties together with exemplary usage, and report on validation methods. The CCF Ontology graph database and API are used in the HuBMAP portal, HRA Organ Gallery, and other applications that support data queries across multiple, heterogeneous sources.
Additional Resources
- The CCF OWL v2.0.1 is available through the NCBI BioPortal Ontology Browser
- Find the CCF OWL v2.0.1
- A GitHub repository with basic information about working with the ontology can be found at https://github.com/hubmapconsortium/ccf-ontology.
- N3.js (https://www.npmjs.com/package/n3) is used as a graph database.
Tissue registration and exploration user interfaces in support of a human reference atlas
Börner K, Bueckle A, Herr BW 2nd, Cross LE, Quardokus EM, Record EG, Ju Y, Silverstein JC, Browne KM, Jain S, Wasserfall CH, Jorgensen ML, Spraggins JM, Patterson NH, Weber GM
Published in Commun Biol., December 2022.
Abstract
Seventeen international consortia are collaborating on a human reference atlas (HRA), a comprehensive, high-resolution, three-dimensional atlas of all the cells in the healthy human body. Laboratories around the world are collecting tissue specimens from donors varying in sex, age, ethnicity, and body mass index. However, harmonizing tissue data across 25 organs and more than 15 bulk and spatial single-cell assay types poses challenges. Here, we present software tools and user interfaces developed to spatially and semantically annotate ("register") and explore the tissue data and the evolving HRA. A key part of these tools is a common coordinate framework, providing standard terminologies and data structures for describing specimen, biological structure, and spatial data linked to existing ontologies. As of April 22, 2022, the "registration" user interface has been used to harmonize and publish data on 5,909 tissue blocks collected by the Human Biomolecular Atlas Program (HuBMAP), the Stimulating Peripheral Activity to Relieve Conditions program (SPARC), the Human Cell Atlas (HCA), the Kidney Precision Medicine Project (KPMP), and the Genotype Tissue Expression project (GTEx). Further, 5,856 tissue sections were derived from 506 HuBMAP tissue blocks. The second "exploration" user interface enables consortia to evaluate data quality, explore tissue data spatially within the context of the HRA, and guide data acquisition. A companion website is at https://cns-iu.github.io/HRA-supporting-information/ .
Additional Resources
- All existing ASCT+B tables can be downloaded from https://humanatlas.io/asctb-tables.
- Organs in the CCF-HRA 3D Reference Object Library are available as GLB files at https://humanatlas.io/3d-reference-library?version=1.4.
- Further, these GLB files are semantically tagged and linked into the CCF-HRA Ontology (CCF.OWL) via the JSON-LD data format.
Corresponding Authors
Katy Börner and Andreas Bueckle
Grant support
NIH Award Nos. OT2 OD026671, U54 AI142766, U54 DK120058, U54 HL145608, and OT2 OD026675
Annotation of Spatially Resolved Single-cell Data with STELLAR
Brbić M, Cao K, Hickey JW, Tan Y, Snyder MP, Nolan GP, Leskovec J.
Published in Nature Methods, November 2022.
Abstract
Accurate cell-type annotation from spatially resolved single cells is crucial to understand functional spatial biology that is the basis of tissue organization. However, current computational methods for annotating spatially resolved single-cell data are typically based on techniques established for dissociated single-cell technologies and thus do not take spatial organization into account. Here we present STELLAR, a geometric deep learning method for cell-type discovery and identification in spatially resolved single-cell datasets. STELLAR automatically assigns cells to cell types present in the annotated reference dataset and discovers novel cell types and cell states. STELLAR transfers annotations across different dissection regions, different tissues and different donors, and learns cell representations that capture higher-order tissue structures. We successfully applied STELLAR to CODEX multiplexed fluorescent microscopy data and multiplexed RNA imaging datasets. Within the Human BioMolecular Atlas Program, STELLAR has annotated 2.6 million spatially resolved single cells with dramatic time savings.
Corresponding Authors
Garry P. Nolan and Jure Leskovec
Contributing groups
Grant support
NIH Award No. U54 HG012723
A user-friendly tool for cloud-based whole slide image segmentation with examples from renal histopathology
Lutnick B, Manthey D, Becker JU, Ginley B, Moos K, Zuckerman JE, Rodrigues L, Gallan AJ, Barisoni L, Alpers CE, Wang XX, Myakala K, Jones BA, Levi M, Kopp JB, Yoshida T, Zee J, Han SS, Jain S, Rosenberg AZ, Jen KY, Sarder P; Kidney Precision Medicine Project.
Published in Commun. Med., August 2022.
Abstract
Background: Image-based machine learning tools hold great promise for clinical applications in pathology research. However, the ideal end-users of these computational tools (e.g., pathologists and biological scientists) often lack the programming experience required for the setup and use of these tools which often rely on the use of command line interfaces.
Methods: We have developed Histo-Cloud, a tool for segmentation of whole slide images (WSIs) that has an easy-to-use graphical user interface. This tool runs a state-of-the-art convolutional neural network (CNN) for segmentation of WSIs in the cloud and allows the extraction of features from segmented regions for further analysis.
Results: By segmenting glomeruli, interstitial fibrosis and tubular atrophy, and vascular structures from renal and non-renal WSIs, we demonstrate the scalability, best practices for transfer learning, and effects of dataset variability. Finally, we demonstrate an application for animal model research, analyzing glomerular features in three murine models.
Conclusions: Histo-Cloud is open source, accessible over the internet, and adaptable for segmentation of any histological structure regardless of stain.
Additional Resources
Corresponding Authors
Pinaki Sarder
Contributing groups
Grant support
NIH Award No. U54 HL145608