HuBMAP unveils new tools, additional data online and publicly available
HuBMAP unveils new tools, additional data online and publicly available
September 1, 2021
The HuBMAP Consortium has announced its latest data portal update, complete with new data, interactive visualization tools, and additional organs as researchers expand their effort to build a cellular atlas of the human body. HuBMAP’s tools and maps are openly available to the public, and seek to accelerate understanding of human cells and tissues. Since the inaugural data release last September, Consortium collaborators have been expanding the information available to the research community via the Data Portal.
Following ongoing data uploads to the portal, there are now a total of 11 organ types from 58 donors, totalling 494 samples and 512 datasets available. In addition to new data, new tools and functionality make the portal easily searchable, with innovative mapping and visualization features.
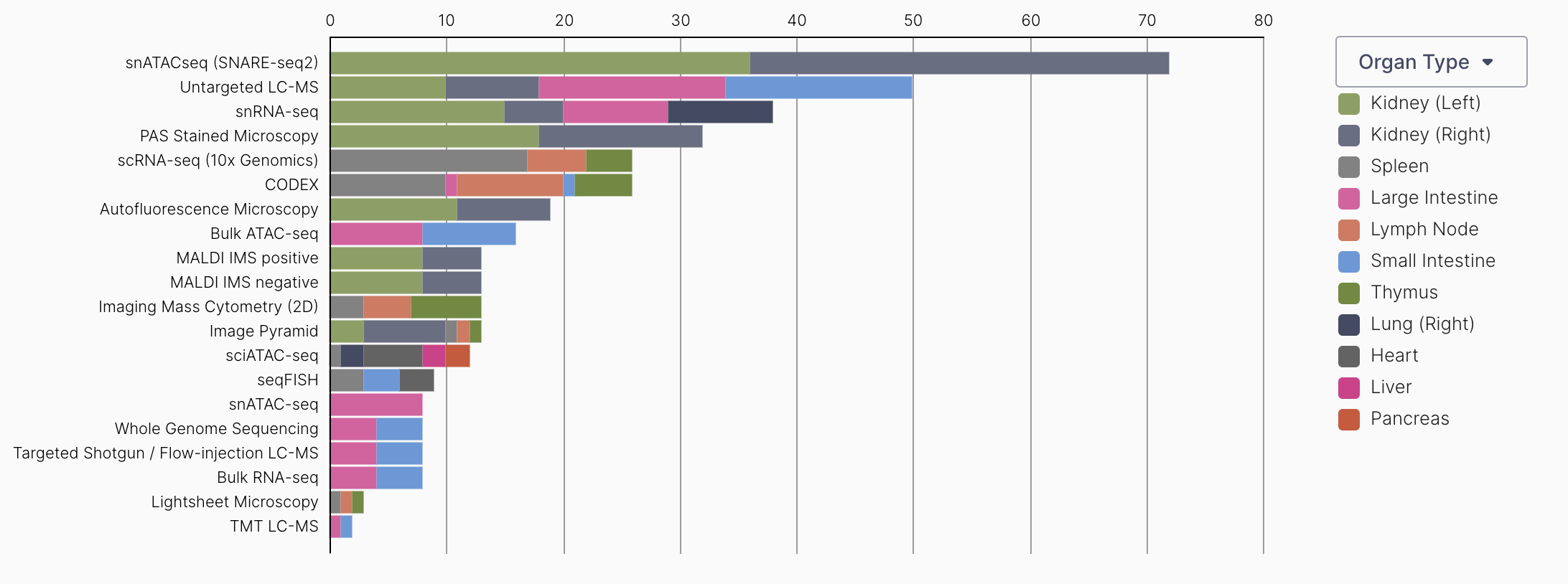
A table of assays by organ type available on the data portal.
Regarding the latest developments from HuBMAP and the data now available, HuBMAP Steering Committee co-chair and PI of the Tissue Mapping Center at Vanderbilt University Jeff Spraggins commented, “Technologies developed by HuBMAP researchers provide data that uniquely span a wide range of spatial scales, from single cells to whole organs, and molecular classes including metabolites, lipids, proteins, and nucleic acids. These data are enabling new possibilities for integration and analysis that will provide a deeper understanding of the molecular drivers of health and disease.”
As part of the Common Coordinate Framework (CCF), users can now browse organ locations through the Exploration User Interface (CCF-EUI) or spatially register a tissue sample with the Registration User Interface (CCF-RUI). The ASCT+B (anatomical structures, cell types, and biomarker) Reporter, another feature long in development by the Consortium and its partners, is a state-of-the-art visualization tool presenting relationships between various anatomical structures and substructures.
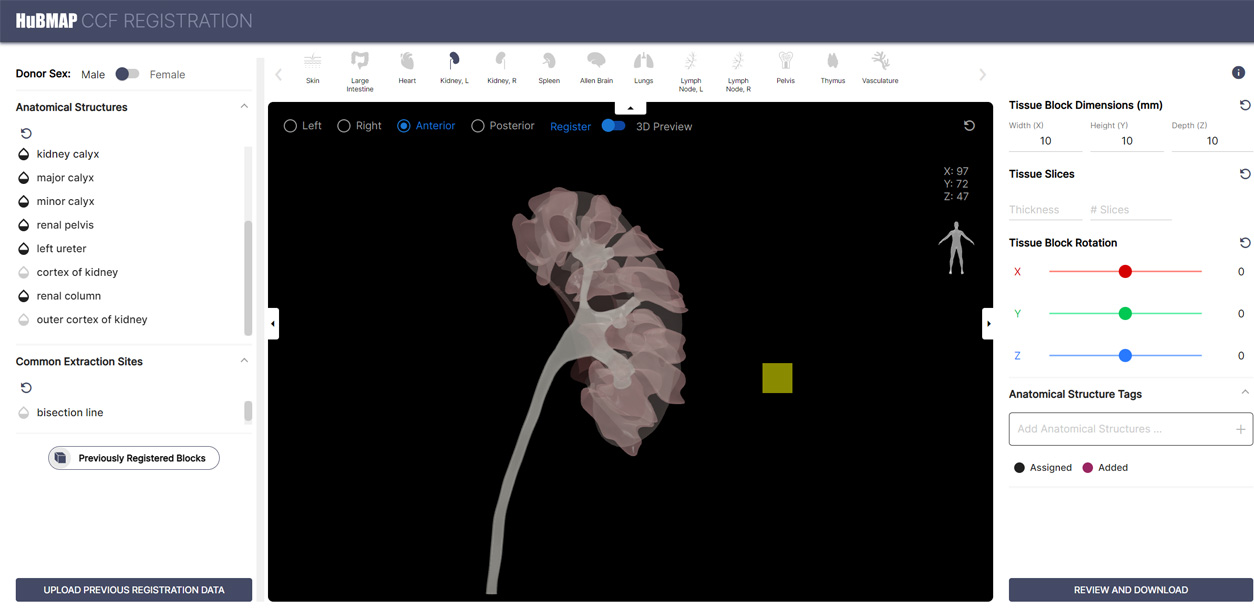
A preview of the CCF RUI.
Two more tools complete the latest round of updates, though some researchers have been taking advantage of these innovations for some time now. A web application that automatically maps scRNA-seq datasets to reference organs, Azimuth leverages a pipeline that inputs a counts matrix of gene expression in single cells, and performs normalization, visualization, cell annotation, and differential expression (biomarker discovery).
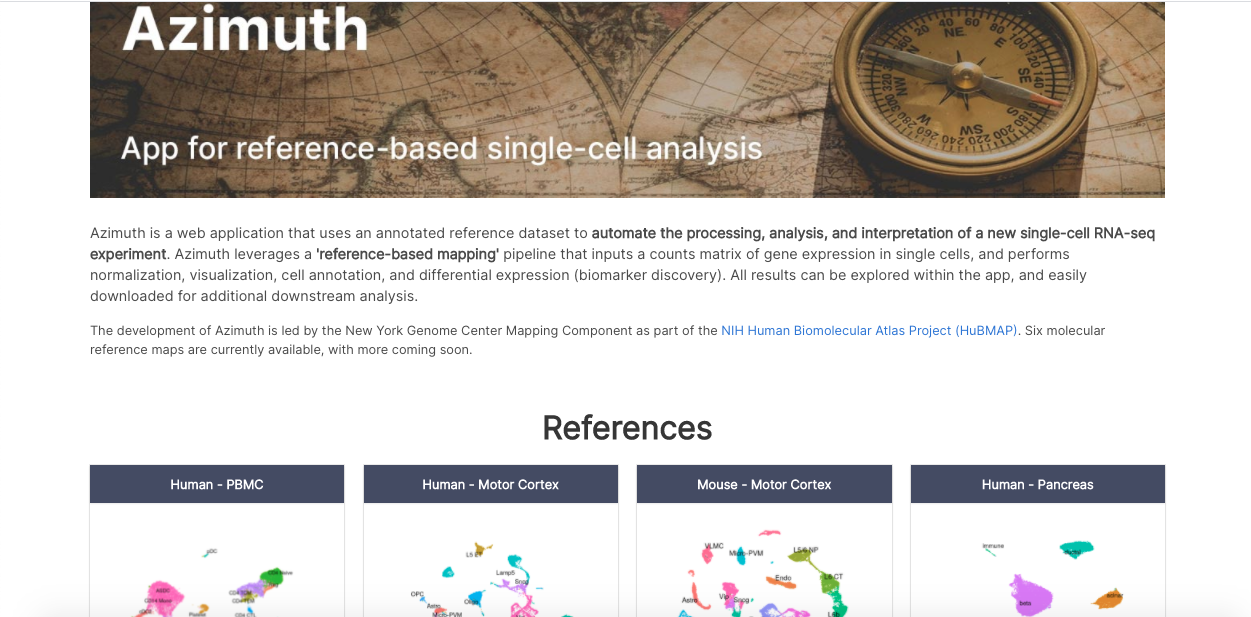
Azimuth homepage
Development efforts over the past year have extended the functionality of the Vitessce visual integration tool for exploration of spatial single cell experiments. Vitessce is designed to support diverse spatial and non-spatial omics and imaging data types and to integrate with a wide range of analysis workflows. Among many other new features, Vitessce now offers support for visualizing 3D volumes via raycasting and direct visualization of annData objects. The portal also provides shareable links to specific states of a Vitessce visualization. Outside the data portal, Vitessce can easily be deployed in other environments, for example, Jupyter Notebooks or standalone websites. There are some datasets on the portal that highlight the capabilities of Vitessce, such as this CODEX dataset and this Lightsheet microscopy dataset. In collaboration with the NYGC Mapping Component, Vitessce visualizations for all Azimuth reference datasets have been made available as well.
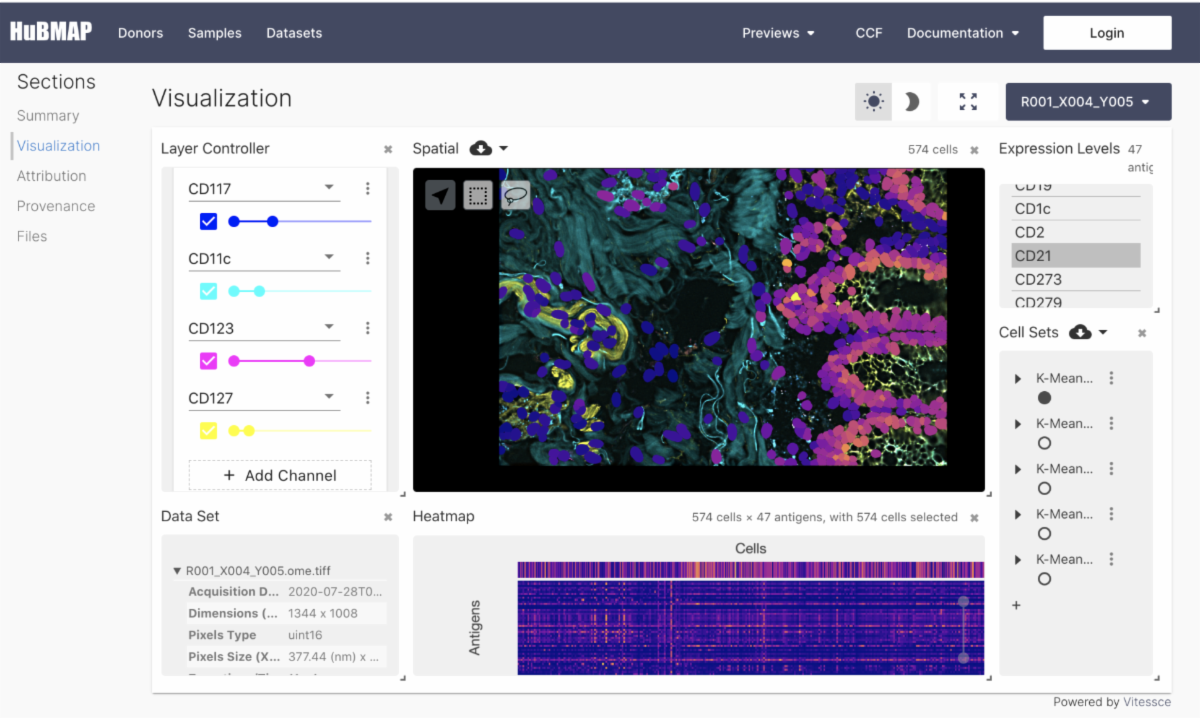
Visualization utilizing Vitessce
Katy Borner, HuBMAP Steering Committee co-chair and PI of the Indiana University HIVE Mapping Component, commented on the Consortium’s excitement surrounding this release and hopes for the future: “The construction of a computable Human Reference Atlas requires close collaboration of experts from engineering and science departments, different institutions and countries. HuBMAP is proud to partner with 15 international consortia (see joint paper) and we look forward to expanding the number and diversity of tissue data used to understand healthy aging in women and men.”
The Consortium and its members will continue to enhance the data portal to ensure its accessibility and provide innovative tools for researchers within and beyond the HuBMAP community, as the available data continues to evolve. Future releases will include expanded tools, new organs, and additional datasets.
The availability of new HuBMAP tools and data was made possible by the collaborative efforts across HuBMAP Consortium including:
- Common Coordinate Framework, Exploration User Interface, and Registration User Interface development by HuBMAP HIVE Indiana University Mapping Component, led by Katy Borner.
- Azimuth creation and development by HuBMAP HIVE New York Genome Center Mapping Component led by Raul Satija
- Vitesse creation and development by HuBMAP HIVE Harvard Medical School Tools Component led by Nils Gehlenborg
- Data processing and pipeline development by HuBMAP HIVE Carnegie Mellon University Tools Component, led by Ziv Bar-Joseph
- Data ingest, data management, infrastructure, and project coordination and communication provided by HuBMAP HIVE Pittsburgh Supercomputing Center/University of Pittsburgh Infrastructure Component led by Phil Blood and Jonathan Silverstein
- HuBMAP data is provided by Tissue Mapping Centers, Transformative Technology Development, and Rapid Technology Implementation teams. Learn more about these teams and the research specialties.
Return to Home.
