HuBMAP Tools
The focus of HuBMAP is understanding the intrinsic intra-, inter-, and extra- cellular biomolecular distribution in human tissue. Along with providing tissues for study in the HuBMAP data portal, we have also been developing tools for public use, as well as for our data providers.
HuBMAP tools are described below.
You can read more about HuBMAP data or you can visit the HuBMAP Data Portal.
Human Reference Atlas
A Human Reference Atlas (HRA) is a comprehensive, high-resolution, three-dimensional atlas of all the cells in the healthy human body. The HRA provides standard terminologies and data structures for describing specimens, biological structures, and spatial positions linked to existing ontologies.
Tools in the HRA include the Anatomical Structures, Cell Types and Biomarkers (ASCT+B) Reporter, Cell Population Graphs, the Registration User Interface, the Exploration User Interface, the Organ Gallery in VR, Millitome, and the relevant APIs.
ASCT+B Reporter
A visualization tool for human organ experts to explore and compare Anatomical Structures, Cell Types, and Biomarkers (ASCT+B) Tables
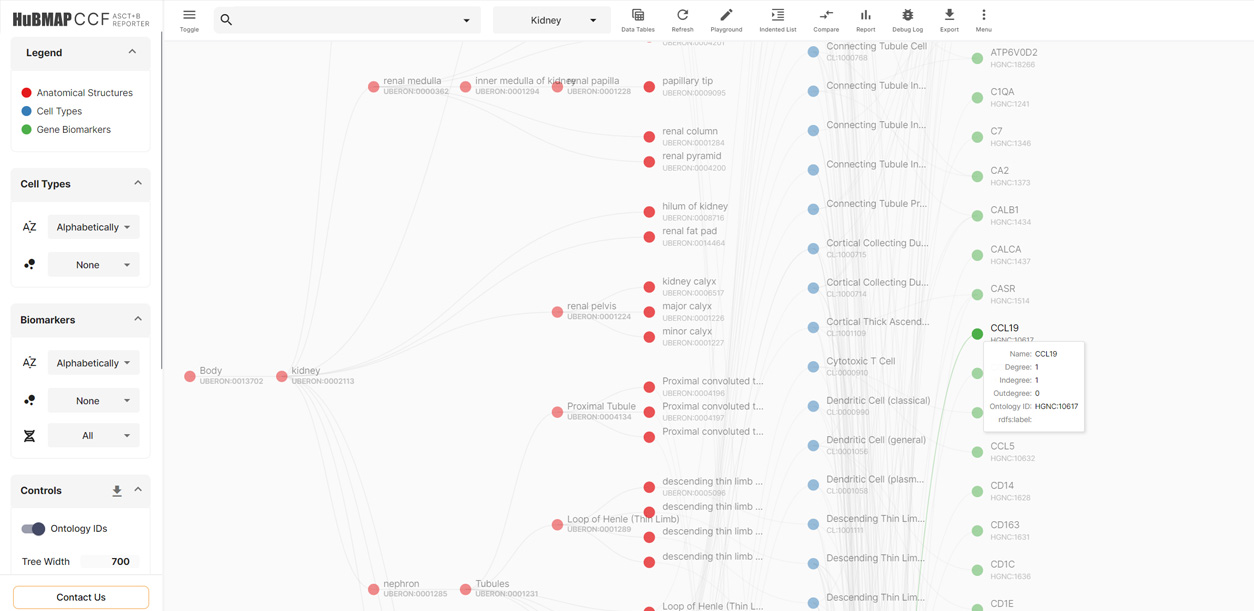
Cell Population Graphs
An interactive tool for exploring and comparing cell populations.
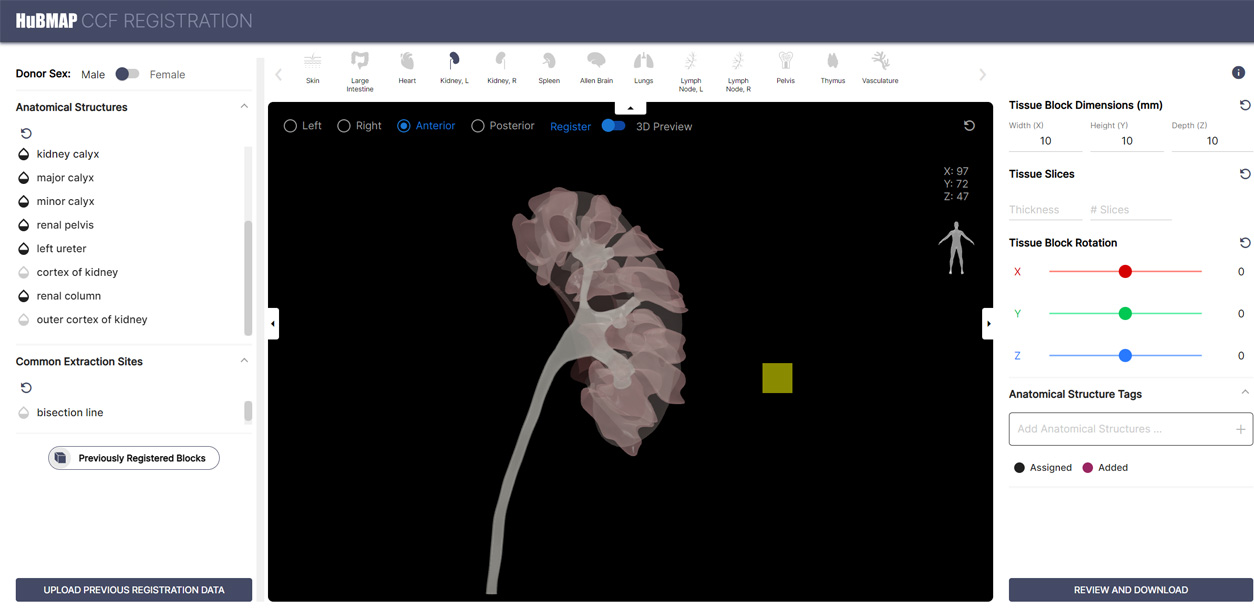
Registration User Interface
A interactive tool for registering tissue blocks spatially and annotating them semantically using ASCT+B Table terms
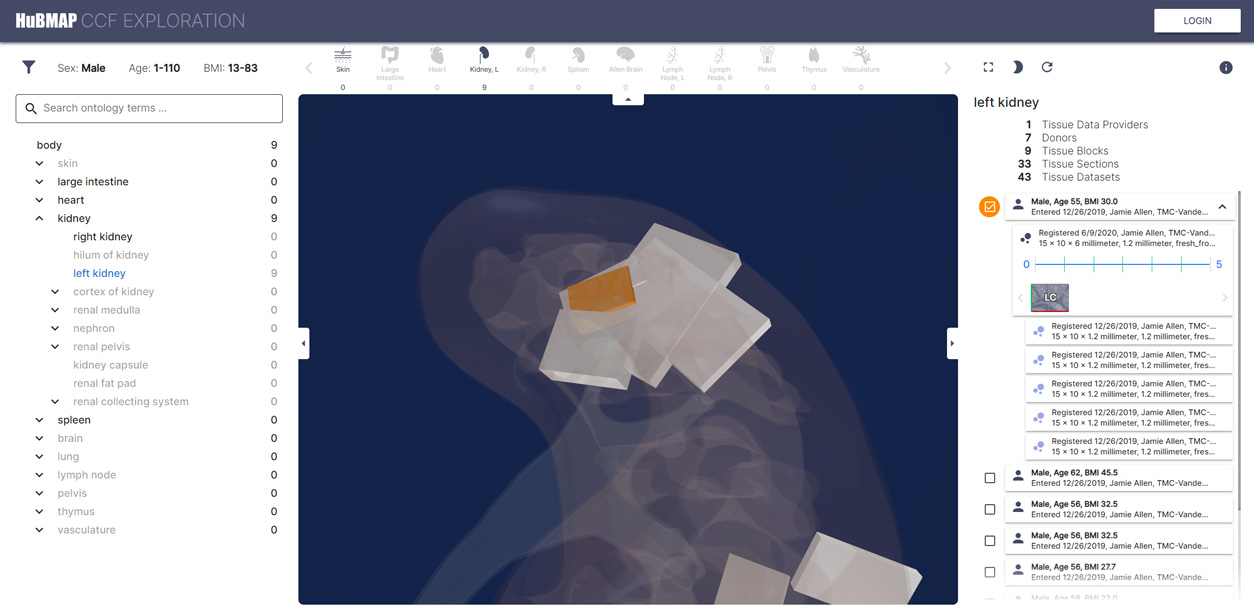
Exploration User Interface
An interactive tool for exploring and validating spatially registered tissue blocks and cell-type populations
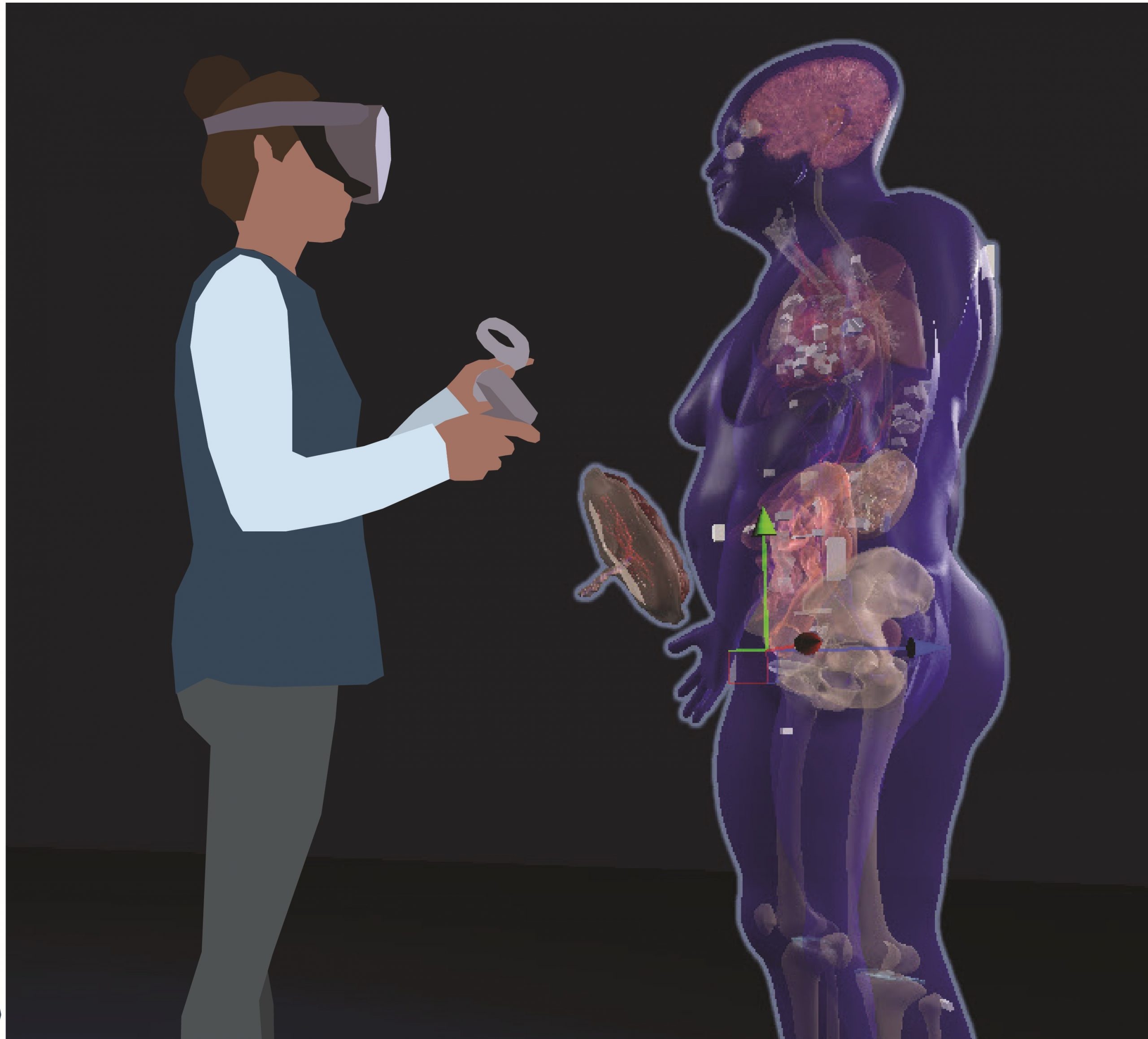
Organ Gallery in VR
An immersive application that allows users to explore 3D reference organs, anatomical structures, and cell types in virtual reality
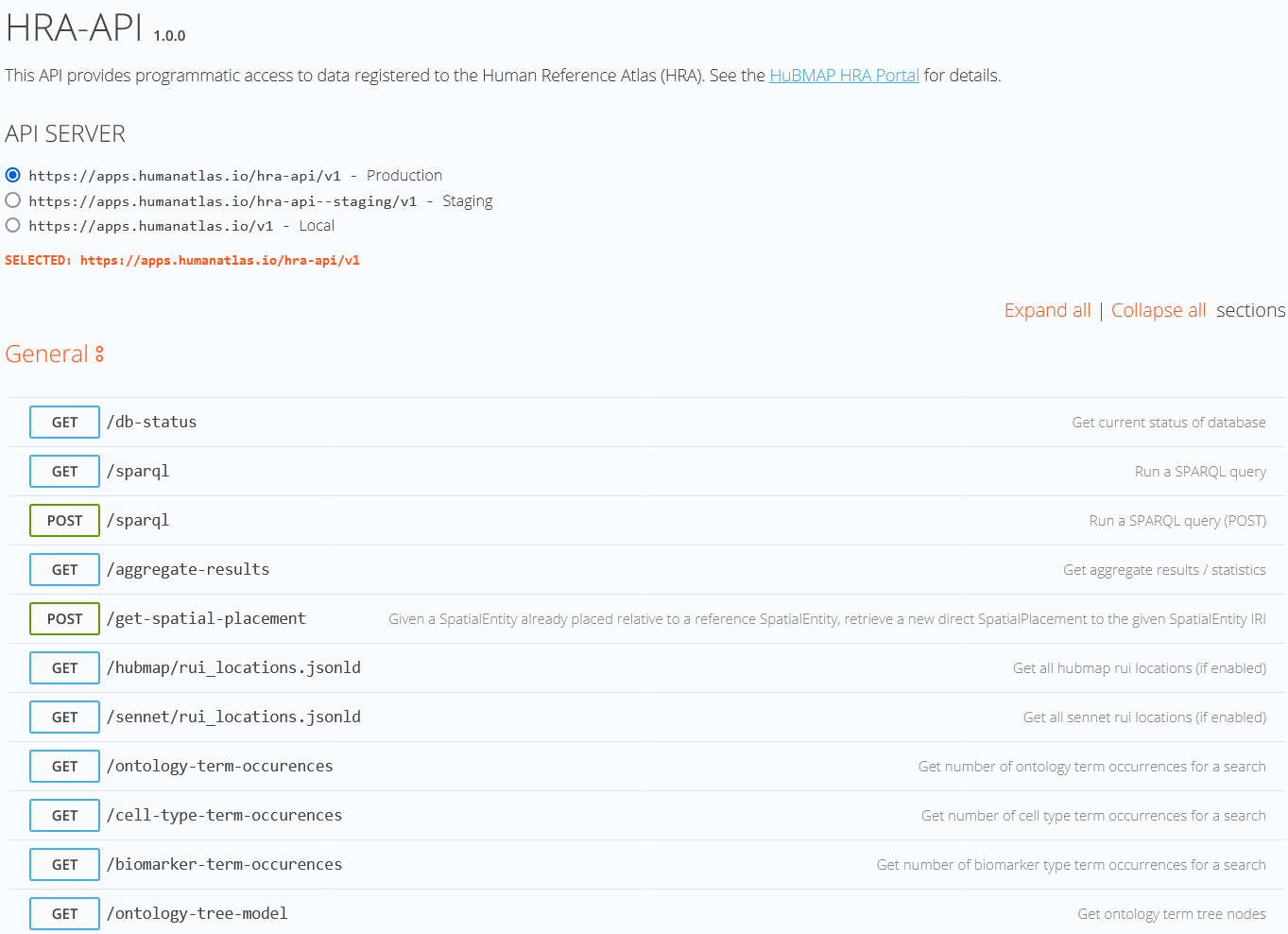
Application Programming Interfaces
Query and interact with the HRA using Python, JavaScript, SPARQL, REST, and more
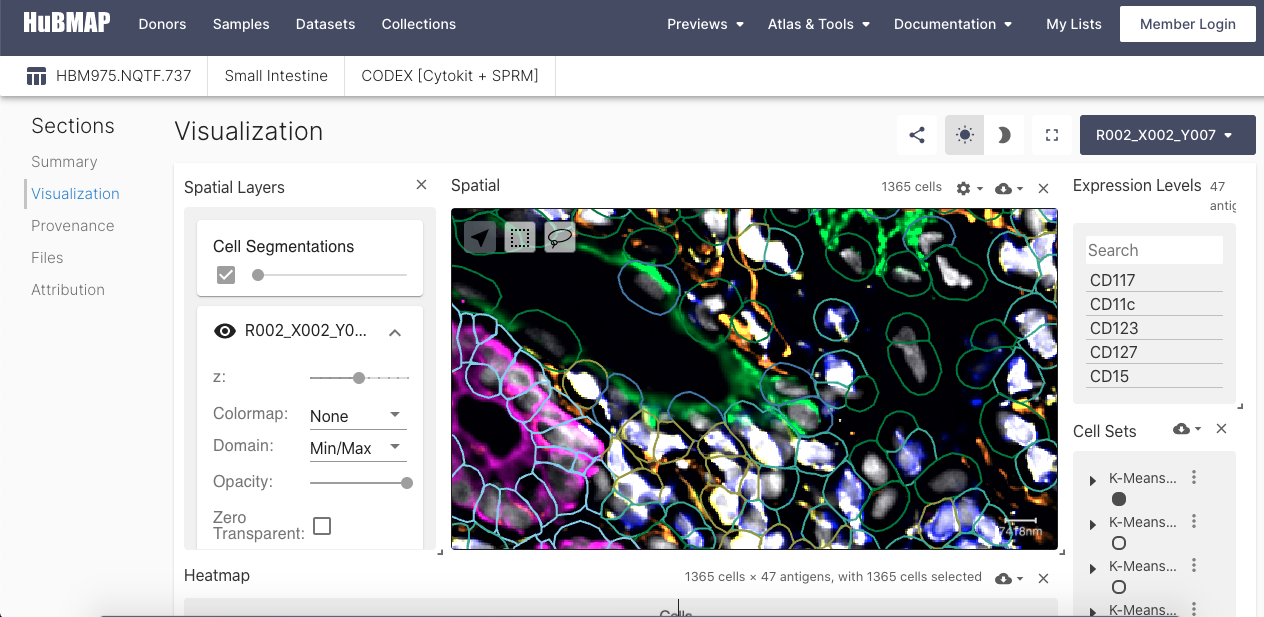
Vitessce
Vitessce enables the exploration of spatially-resolved, integrated single-cell datasets, with reusable interactive components such as a scatterplot, spatial+imaging plot, genome browser tracks, statistical plots, and controller components. An interactive visualization tool, Vitessce is available for datasets that have been processed through HuBMAP HIVE computing hardware and pipelines.
Azimuth is a web application that uses an annotated reference dataset to automate the processing, analysis, and interpretation of a new single-cell RNA-seq experiment. Azimuth leverages a 'reference-based mapping' pipeline that inputs a counts matrix of gene expression in single cells, and performs normalization, visualization, cell annotation, and differential expression (biomarker discovery). All results can be explored within the app, and easily downloaded for additional downstream analysis.
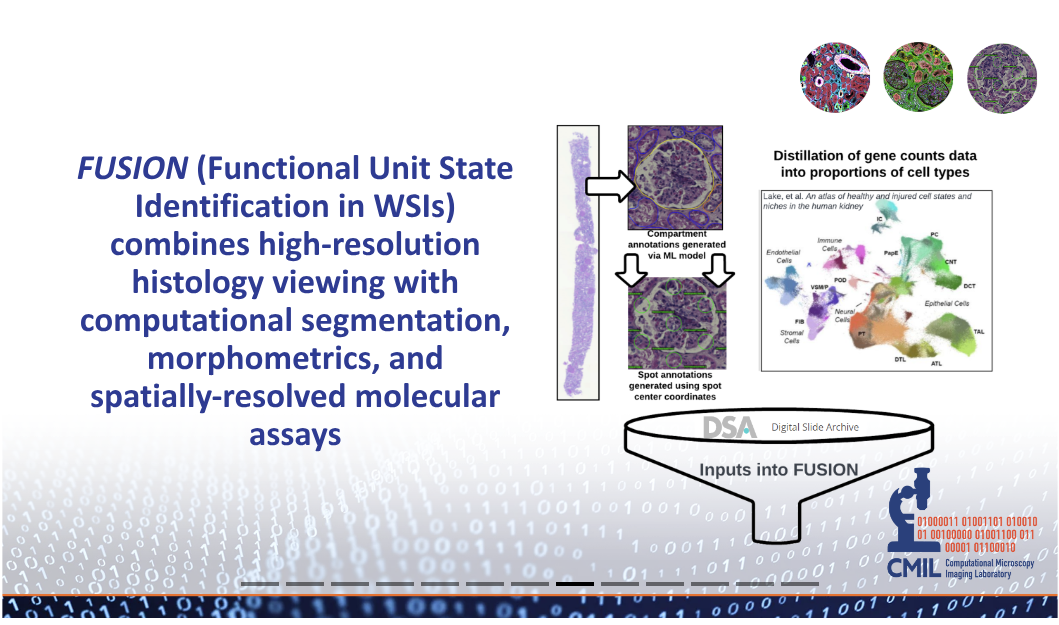
FUSION is an interactive interface to view spatial transcriptomics data integrated with histopathology, driven by artificial intelligence (AI). AI segments Functional Tissue Units (FTU) and links these with gene expression data that informs on major and minor healthy and injured cell types directly mapped on histological features on the FTUs in the WSI.