Human BioMolecular Atlas Program: Scientists Announce New Version of 3D Human Reference Atlas (HRA)
Human BioMolecular Atlas Program: Scientists Announce New Version of 3D Human Reference Atlas (HRA)
March 14, 2025
Coverage and Quality of the Human Reference Atlas (HRA): Experts from many fields of science around the globe have constructed and use the HRA; the HRA Dashboard shows the size and coverage of the HRA data, including the number and type of HRA usage over time, publication, and experimental data, as well as data about atlas donors, users, and authors that will help ensure consistent results and medical relevance for patients. The HRA Dashboard can be explored at https://apps.humanatlas.io/dashboard.
A new paper in the prestigious journal Nature Methods describes the latest expansion of the Human Reference Atlas (HRA) and its Common Coordinate Framework (CCF), a central set of 3D tools for mapping the human body at the smallest scales created by the Human BioMolecular Atlas Program (HuBMAP) funded by the NIH Common Fund. The “Resource” paper, which appeared on March 13, 2025, shares Findable, Accessible, Interoperable, Reusable (FAIR) data and open code for the 3D HRA for the scientific community to use.
“It takes a village of bold interdisciplinary innovators to map the human body at single-cell resolution,” said Katy Börner, principal investigator of the HuBMAP Mapping Component at Indiana University. “More than 50 different algorithms and thousands of datasets need to be harmonized to arrive at a multiscale atlas that is more than the sum of parts.”
The HRA provides standard terminologies and data structures for describing specimens, biological structures, and spatial positions linked to existing ontologies, and uses data from NIH Common Fund programs, including the Human Biomolecular Atlas Program (HuBMAP), Cellular Senescence Network (SenNet) Genotype-Tissue Expression project (GTEx), and other data portals. A flexible hybrid cloud microservices architecture developed by the Pittsburgh Supercomputing Center team led by Phil Blood and the University of Pittsburgh team led by Jonathan Silverstein is used to provide efficient and sustainable storage and access to data, plug-and-play integration of 50+ algorithms, and access to computational resources co-located with data and tools at no cost to the research community. The CMU Tools Component team, led by Matt Ruffalo and Sarah Teichman, is responsible for algorithm integration and validation. New data is mapped to the HRA using Azimuth1 developed by the NYGC Mapping Center (MC) led by Rahul Satija and John Marioni, and spatially using methods2,3 and HRA user interfaces4 developed by Börner’s team.
“The HRA takes on the incredibly important tasks to organize and make increasingly complex molecular and cellular biological data accessible to all levels of education and expertise from students of human biology to basic, clinical and translational scientists and to the general public,” said Gloria Pryhuber, principal investigator of the HuBMAP-Lung project at the University of Rochester.
Experts from 26 consortia are recognized for contributing to the construction of the HRA with funding and strong support by the National Institutes of Health (NIH), the Chan Zuckerberg Initiative (CZI), and the international Human Cell Atlas (HCA) effort:
- Allen Brain Atlas
- BRAIN Initiative Cell Atlas Network (BICAN) and BRAIN Initiative Cell Census Network (BICCN)
- Cellular Senescence Network (SenNet)
- Chan Zuckerberg Initiative (CZI) Seed Networks for HCA
- CIFAR MacMillan Multiscale Human Program
- Common Fund Data Ecosystem (CFDE) Data Distillery
- CZI Pediatric Networks for HCA
- Developmental Genotype-Tissue Expression (dGTEx)
- Extracellular RNA Communication Consortium (ERCC)
- GenitoUrinary Developmental Molecular Anatomy Project (GUDMAP)
- Genotype-Tissue Expression project (GTEx)
- Helmsley Gut Cell Atlas
- Horizon 2020 funding for the HCA
- Human Biomolecular Atlas Program (HuBMAP)
- Human Tumor Atlas Network (HTAN)
- Kidney Precision Medicine Project (KPMP)
- Molecular Atlas of Lung Development Program (LungMAP)
- Oklahoma Medical Research Foundation (OMRF)
- Pediatric Center of Excellence in Nephrology (PCEN) at Washington University in St. Louis
- (Re)building a Kidney (RBK)
- Stimulating Peripheral Activity to Relieve Conditions (SPARC)
- The Cancer Genome Atlas (TCGA)
- The Knockout Mouse Project (KOMP)
- United Kingdom Research and Innovation Medical Research Council funding for the HCA
- Wellcome funding for HCA pilot projects
- 4D Nucleome (4DN)
“Building the HRA offers interdisciplinary opportunities for all career stages,” said Andreas Bueckle, Research Lead in the HuBMAP Mapping Component at Indiana University. “I enjoy harmonizing and visualizing datasets from the whole-body to subcellular structures within a 3D common coordinate framework and virtual reality with my junior colleagues.”
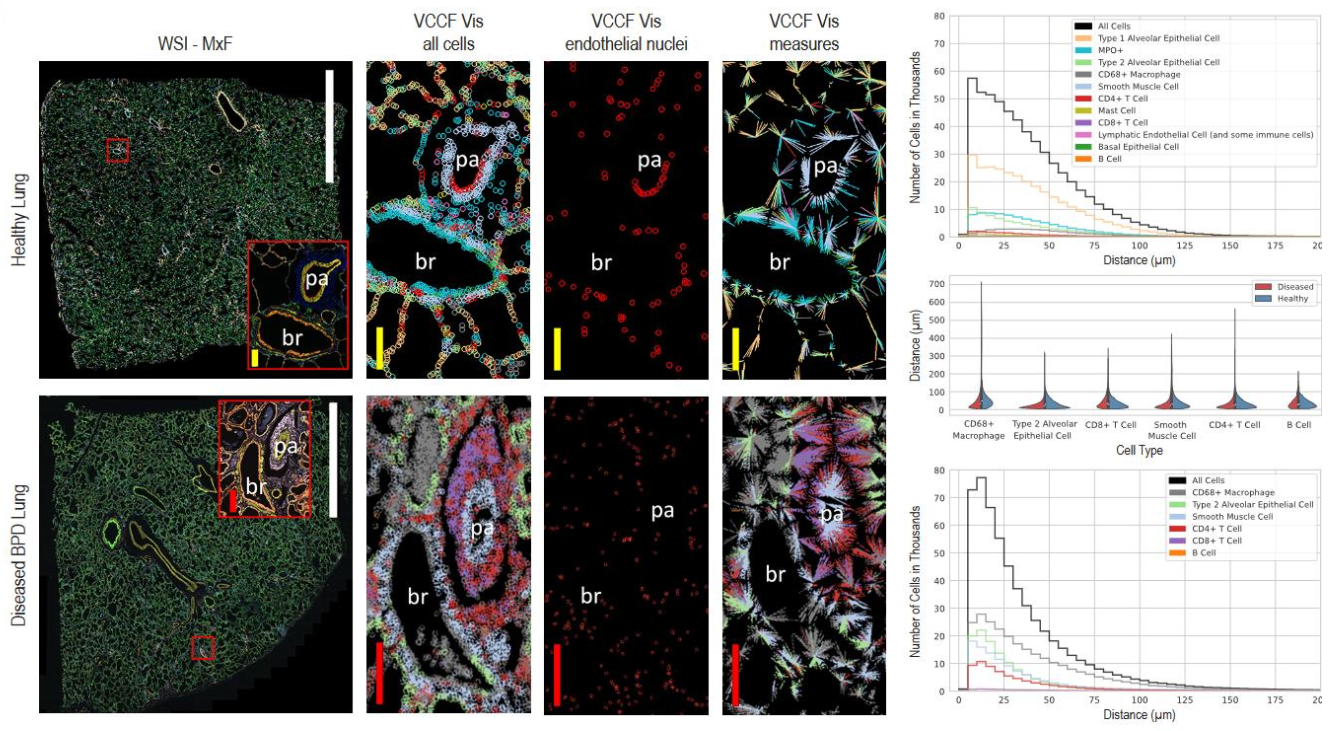
The new paper describes the stories from scientific users of the HRA and lessons learned in making the most productive use of the data and tools underlying it. The report also updates terminology and data formats to ensure scientists are making “apples to apples” comparisons. Finally, it previews ways of using the tools that comprise the HRA to explore new questions about human health and disease. Specifically, the paper showcases how healthy lung reference data can be compared with diseased data using spatial cell distance distribution patterns computed for single cell multiplexed immunofluorescence microscopy images, see below figure for a comparison of healthy vs. diseased lung data. For colon data, it demonstrates how multi-level cell neighborhoods can be computed for multiplexed CODEX image data to study the structure of functional tissue units such as intestinal crypts or immune follicles.
“Rapid advances in single-cell-omics have transformed our understanding of biology, generating vast data that no single group can fully analyze,” said John Hickey, principal investigator at Duke University. “The HRA unifies these datasets to empower spatial analyses across scales and tissues, enabling new questions about cell organization and breakthrough discoveries.”
HuBMAP, funded by the Common Fund, in the Office of the Director of the U.S. National Institutes of Health, aims to construct a reference 3D structural, cellular, and molecular atlas of the healthy adult human body. The HuBMAP Data Portal serves experimental datasets and supports data processing, search, filtering, and visualization. The Human Reference Atlas (HRA) Portal provides open access to atlas data, code, procedures, and instructional materials.
“When HuBMAP launched in 2018, bringing together so many collaborators with different research priorities, it was hard to imagine we’d reach this point,” said Robin Flaus Scibek, HuBMAP coinvestigator at PSC. “Thanks to the IU team’s leadership and dedication, we now have a resource that integrates efforts across 26 different consortia. I’m excited to see how these tools drive future scientific breakthroughs and open new doors for discovery.”
A central goal of the effort to create a 3D map of the human body down to microscopic scales is to understand how organs and tissues in the body work. This information will drive a better understanding of what constitutes a healthy human, as well as the changes that drive aging, disease, and other disorders, with the aspiration of providing doctors with targets for better treatments for the range of human illness.
“The HRA strives to provide a reference that is both comprehensive and maximally reusable,” said Bruce W. Herr II, HRA senior software architect and project manager at Indiana University. “The Linked Open Data architecture coupled with world class algorithms, high quality data, and subject matter expertise helps provide a unique resource for the community.”
The 7th release of the HRA v2.0 covers:
- 36 organs with 4,499 unique anatomical structures
- 1,195 cell types
- 2,089 biomarkers (for example genes, proteins, or lipids) linked to human physical development as well as reference structures in the body that can be used as waypoints in “navigating” through the body at a microscopic scale
The HRA allows scientists to map their own data accurately into the body’s structure in three ways:
- Software annotation tools allow researchers to attach non-spatial data and information to specific anatomic points in the body, keeping those data easily available for analysis of those structures.
- Azimuth leverages a 'reference-based mapping' pipeline to normalize and visualize single-cell RNA-seq or ATAC-seq data, perform cell annotation and biomarker discovery.
- Validated antibody panels use antibody proteins available to scientists either from commercial vendors or from other scientists to identify molecules important to specific life functions and their precise locations in the body.
- Spatial tissue registration allows researchers to fix the sources of their data in the 3D map with a common coordinate system, much like a GPS gives coordinates for a location on Earth.
Future goals for the HRA include further development to make it and its tools more accurate and useful to scientists. HuBMAP and its collaborators will expand and update its reference data, antibody panels, and list of organs supported. They will also improve and expand the number of web-based tools and APIs to improve access for scientists. Finally, they will expand the connections between the HRA Portal and the HuBMAP, SenNet and other Data Portals to improve construction and usage of the HRA.
“It has been exciting building the flexible hybrid cloud infrastructure that makes data supporting the HRA readily and freely available to anyone,” said PSC’s Blood. “This approach will facilitate sustained access to these valuable datasets beyond the end of the programs that initially funded them.”
Community input to HRA user stories, data, code, user interfaces, and training materials is welcome. Experts interested to learn more about or contribute to the HRA effort are encouraged to register for the monthly HRA working group events online, at https://iu.co1.qualtrics.com/jfe/form/SV_bpaBhIr8XfdiNRH.
The Human BioMolecular Atlas Program (HuBMAP) is funded by the National Institutes of Health Common Fund. Award Number: OT2OD033759.
References
- Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573-3587.e29 (2021).
- Börner, K. et al. Anatomical structures, cell types and biomarkers of the Human Reference Atlas. Nat. Cell Biol. 23, 1117–1128 (2021).
- Quardokus, E. M. et al. Organ Mapping Antibody Panels: a community resource for standardized multiplexed tissue imaging. Nat. Methods 20, 1174–1178 (2023).
- Börner, K. et al. Tissue registration and exploration user interfaces in support of a human reference atlas. Commun. Biol. 5, 1369 (2022).
Contact:
Ken Chiacchia
Pittsburgh Supercomputing Center
412-268-5869
chiacchi@psc.edu
Robin Flaus Scibek
Pittsburgh Supercomputing Center
412-268-1326
flaus@psc.edu