Pancreatic Lymph System, Healthy Pregnancy Foci of HuBMAP Collaborations
Pancreatic Lymph System, Healthy Pregnancy Foci of HuBMAP Collaborations
HuBMAP SciTech Webinar, February 14, 2022
Projects to elucidate the anatomy of the pancreas and the functions of its associated lymphatic system, as well as the healthy pregnant uterus and placenta, were the focus of two presentations by HuBMAP collaborators at the February Webinar. The University of Florida (UF) TMC, Pacific Northwest National Laboratory (PNNL) TMC and Texas Advanced Computing Center (TACC) have joined to explore how the lymph system interacts with the pancreas; collaborators at the University of California San Diego (UCSD) TMC and the Indiana University (IU) HIVE are investigating reproductive tissues in the course of healthy pregnancy.
Integrative multi-modal molecular imaging of the pancreas and regional lymphatic system
Clay Matthews of the UF TMC presented his group’s work, in collaboration with the PNNL TMC led by Clive Wasserfall as well as TACC, to create the first molecular-level 3D map of how the lymphatic system in the pancreas drains and functions.
The lymphatic system is key to immune surveillance in parts of the body that interact with the environment, Matthews said. The pancreas’s role in digestion gives it a window to the microbiome of the GI tract, potentially providing a pathway for immune cells monitoring the gut for new pathogens as well as pathogenic developments in otherwise commensal microbes. Communication between pancreatic cells and immune cells, as well as the routes the former take through and between pancreatic and regional draining lymph nodes, may establish molecular-level relationships that will have biological and medical ramifications.
Matthews took the audience through the processing steps for the single pancreas analyzed to date, separating the organ into 21 blocks running from the head through the tail of the organ, with the slabs in turn split into quadrants. In addition to separating the organ in tissues with distinct functional roles, this process also enables elucidation of connections between the organ and specific lymphatic ducts leading to a variety of lymph nodes. For one example, pancreatic endocrine tumors most often initiate in the tail, with consequences for metastatic spread via lymphatic draining and recirculation back into the organ.
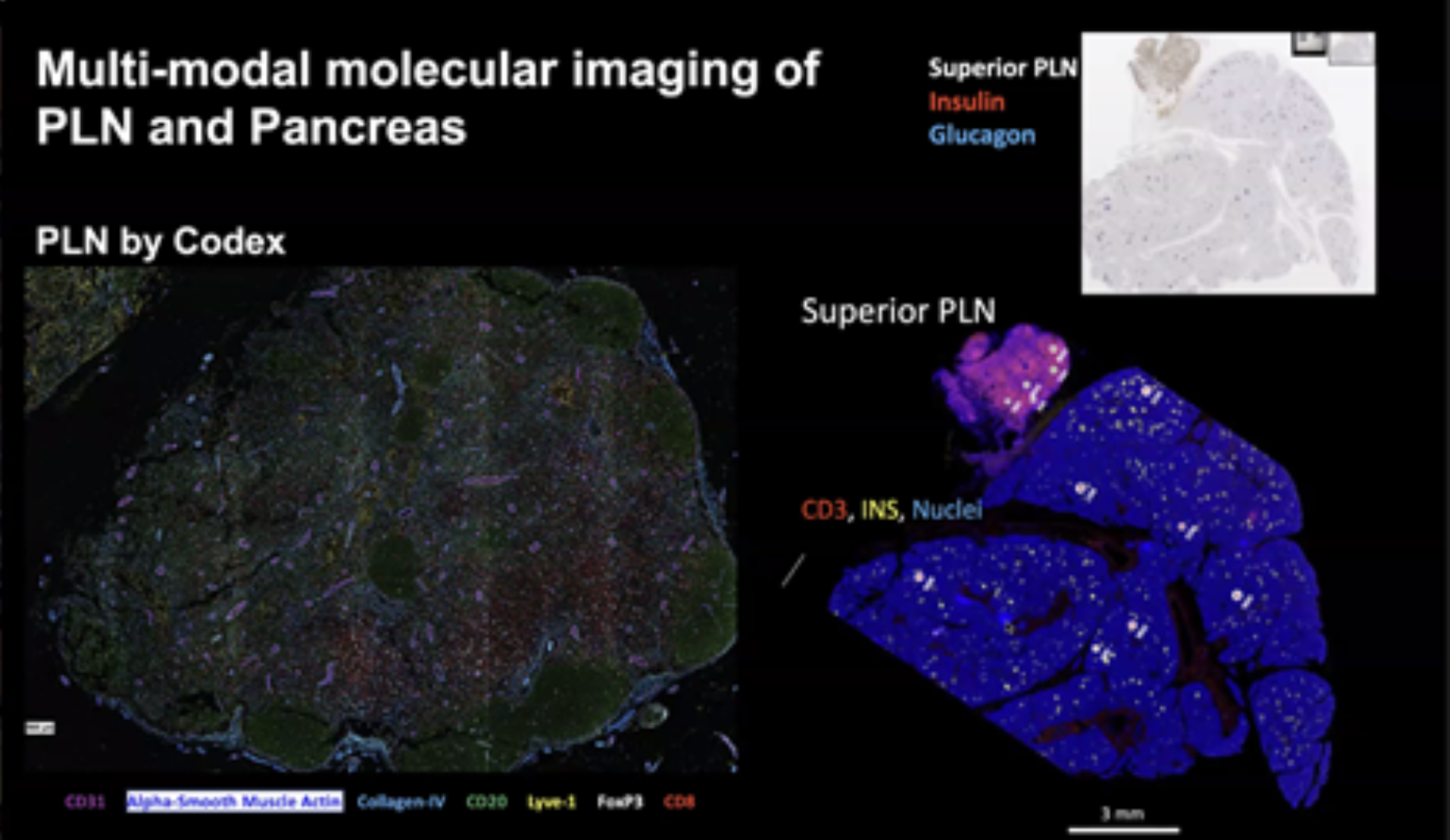
The group is using antibody-based and RNA barcoding reagents to characterize gene expression in the tissues using formalin-fixed, paraffin-embedded (FFPE) tissues to illuminate regions of interest as small as single cells. The result is a rich, region-by-region dataset characterizing biological function. The work employs the GeoMx digital spatial profiler as well as CODEX multiplexed imaging. An initial sequencing data array is currently being analyzed, with a primary analysis of each assay, head-to-head comparisons of orthogonal assays and integration of MASS spec and GeoMx data planned for this spring. The long-term plan will involve placing the data on 3D maps using a common coordinated framework and the TACC/U Texas 3D model of the pancreas.
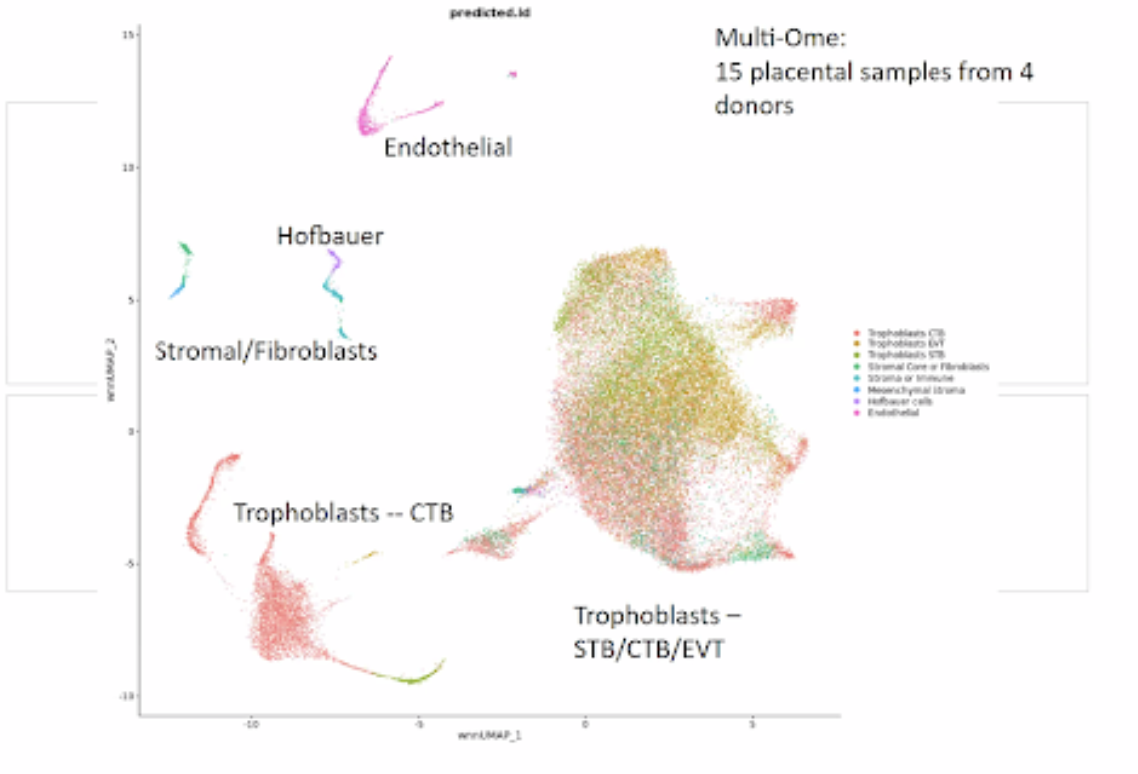
Building 3D tissue maps for the placenta and pregnant uterus
Louise Laurent of the UCSD TMC described her group’s collaboration with the IU HIVE, led by Katy Börner, to map the cell-level anatomy of the placenta and uterus in pregnancy.
Placental tissue is layered, with fetal-derived tissue in the umbilical cord, a mix of fetal and maternal tissues in three membranous layers, and a mix of fetal and maternal tissues in the disc layer. MRI and MRI angiography provide detailed images, but the latter in particular are affected by artifacts that will need to be corrected in the final 3D model.
The group is recruiting healthy pregnant women without comorbidities or complications and no history of poor pregnancy outcomes. The team plans to enroll 50% of subjects with a planned vaginal delivery and 50% with a planned C-section and tubal sterilization. In addition, 50% will be selected each for female vs. male fetuses; white vs. non-white; and Hispanic vs. non-Hispanic. Recruitment is roughly half complete, with the group currently collecting tissues and imaging data (first and second trimester ultrasound and third trimester MRI).
Sampling of the placenta is undertaken at two sites in each organ, with tissues split between fetal, central, and maternal tissues and each analyzed for RNA and self-labeling protein tagging (SNAP-tag) of each sample divided into six sections. The group is also sampling anatomically distinct portions of the fallopian tubes and uterine endomyometrium as well. They will analyze mRNA transcripts (RNAseq), transposase-accessible areas of the chromatin/DNA (ATACseq) and extracellular matrix markers in the three organ types. They are currently working to improve resolution of Visium Spatial Gene Expression imaging to enable visualization of the blood vessels.
An important facet of the work will involve integration of the collaboration’s results with the CCF Organ VR Gallery, which will enable investigators to explore the anatomy via the Oculus VR environment. The gallery code can be seen at https://github.com/andreasbueckle/ccf-organ-vr-gallery, with a demo at https://www.youtube.com/watch?v=S9pBOlSfsnc and a preprint of the work at https://osf.io/z9gm3/. The collaborators are also participating in the current HuBMAP RUI registration contest, with final tissue blocks and samples due by March 4. The RUI tutorial can be found at https://youtu.be/gY3_-LIoKaU, and its SOPs at https://doi.org/10.5281/zenodo.5575776; questions about the contest should be directed to abueckle@iu.edu.
Return to Home.
