Human Reference Atlas Charts Human Body at Single Cell Level
Human Reference Atlas Charts Human Body at Single Cell Level
Will allow computer-guided exploration of human health and disease to identify new treatments
November 8, 2021
An international collaboration of scientists has created the first draft of a digital atlas of the 3D structure of the human body at the level of individual cells. The Human Reference Atlas (HRA) charts the human body in the finest detail, helping scientists and doctors to explore how the smallest structures in the body contribute to its function in health and disease; compare how disease processes affect the functions of cells and tissues; and identify treatment targets for diseases such as cancer, diabetes, heart disease, kidney disease and viral and bacterial infection.
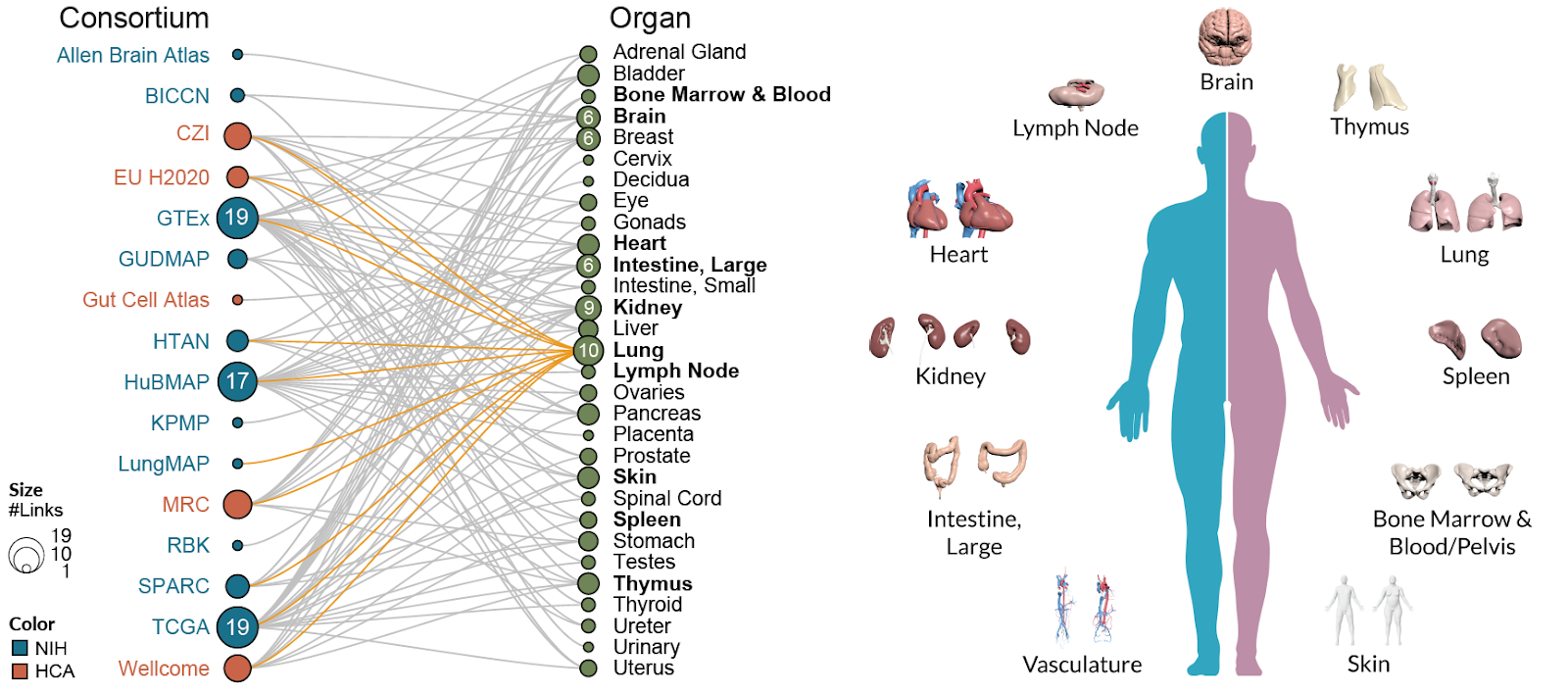
The HRA has been built by a collaboration of 16 international scientific consortia, which reported its results in the prestigious scientific journal Nature Cell Biology today.
“It takes a village of organ experts, single-cell experts, computer scientists, and engineers to construct an open, computable, and authoritative reference atlas of the healthy human body,” said Katy Börner, Victor H. Yngve Distinguished Professor of Engineering & Information Science at the Luddy School of Informatics, Computing, and Engineering at Indiana University (IU), principal investigator of The Human Body Atlas project at IU, and the corresponding author of the paper. “It is exciting to see this interactive atlas come into existence and to explore its first-use cases.”
Börner’s Human Body Atlas project is a part of the U.S. National Institutes of Health Common Fund’s Human Biomolecular Atlas Program (HuBMAP), which involves more than 350 experts from 28 institutions across the world. HuBMAP scientists are working on a kind of “Google Maps” for the body that will allow scientists to explore the body in fine detail with comprehensive, precise, and verified information about specific locales, much like clicking on an address in Google Maps.
Today’s technology allows scientists to read genetic code, tag which genes are and are not active, and identify individual proteins as well as other important biomolecules - all at the level of individual cells. The amount of raw data being generated by these technologies is vast, far more than human brains can understand directly, especially when trying to envision how the biochemical actors play their roles across the body in three dimensions. HuBMAP’s 28 specialized centers are refining, developing, and validating a host of technologies for discovering, communicating, and analyzing this kind of information. By combining their efforts, the centers aim to create a single repository for a massive amount of microscopic and biochemical data that can be searched by computers and visualized interactively so human eyes and minds can make sense of it.
As described in the paper, the HRA’s “Anatomical Structures, Cell Types, plus Biomarkers” (ASCT+B) framework provides a “look-up table,” or index, for a number of 3D maps of the human body - linking the information at each locale to the unique developmental process and identity of the tissues and cells that make them up -across organs present in the ASCT+B tables. Scientists can use these tables to identify specific cell types and see where in the body those cells are located. As a reference for healthy tissue, the ASCT+B tables can also be used to identify changes in the molecules found in a given cell type in normal aging or disease, helping scientists identify targets for drug or other treatments.
The atlas also includes large amounts of metadata that will help scientists keep track of each bit of information and how it was discovered and verified, easily allowing them to update, correct, or expand as new research is generated.
Interactive user interfaces (UI) are freely available online and can be explored at:
The Visible Human MOOC features demo videos of the HuBMAP portal, data, and tools.
The work was funded, in part, by NIH Awards 1OT2OD026671, 1U54DK120058, 1UH3CA246594, 1U54AI142766, 1UG3CA256960, 1UG3HL145609, 1U54HL145608, 1U54HL145611, 1UH3DK114933, 1DK110814, 1DK107350, 1U54HG010426, 1R01HL137811, 1U01DK107350, 1U24DK110814, 1U01HL148861, 1U01HL148867, the NIDDK Kidney Precision Medicine Project grant 1U2CDK114886, the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), Department of Health and Human Services under BCBB Support Services Contract HHSN316201300006W/HHSN27200002, Helmsley Charitable Trust 2018PG-T1D071 and 2004-0381, the Allen Institute, Wellcome-Sanger WT206194, and the Chan Zuckerberg Initiative CZF2019-002442.
Return to Home.
